Keywords
Ovarian cancer, proteomics, personalized medicine, multiplex immunoassay, tumor marker, recurrence, proximity extension assay
Background: Following first-line treatment, over 80% of advanced ovarian cancer cases suffer recurrence. Treatment of patients with recurrence based on CA125 has not resulted in improvements in outcome postulating that we need biomarkers for earlier detection. A tumor-specific array of serum proteins with advanced proteomic methods could identify personalized marker signatures that detect relapse at a point where early intervention may improve outcome.
Methods: For our discovery phase, we employed the proximity extension assay (PEA) to simultaneously measure 1,104 proteins in 120 longitudinal serum samples (30 ovarian cancer patients). For our validation phase, we used PEAs to concurrently measure 644 proteins (including 21 previously identified candidates, plus CA125 and HE4) in 234 independent, longitudinal serum samples (39 ovarian cancer patients).
Results: We discovered 23 candidate personalized markers (plus CA125 and HE4), in which personalized combinations were informative of recurrence in 92% of patients. In our validation study, 21 candidates were each informative of recurrence in 3-35% of patients. Patient-centric analysis of 644 proteins generated a refined panel of 33 personalized tumor markers (included 18 validated candidates). The panel offered 91% sensitivity for identifying individualized marker combinations that were informative of recurrence.
Conclusion: Tracking individualized combinations of tumor markers may offer high sensitivity for detecting recurrence early and aid in prompt clinical referral to imaging and treatment interventions.
Ovarian cancer, proteomics, personalized medicine, multiplex immunoassay, tumor marker, recurrence, proximity extension assay
High-grade serous carcinoma (HGSC) is the deadliest ovarian cancer (OvCa) subtype, with a 5-year survival rate of 15-30%.1–3 Although most patients achieve remission after first-line treatment, 80-95% of advanced cases will relapse within five years, with median survival of 12-24 months.4–7 With novel therapeutic agents in clinical trials,8,9 there is an urgent need to develop sensitive tools for detecting recurrence to aid in finding the key time for therapeutic intervention.
There is no consensus amongst practice guidelines on the threshold for the kinetics of the classical OvCa biomarker, cancer antigen 125 (CA125), which would signify risk of recurrence when the value is low, particuarly in the small subset of patients where CA125 is not highly elevated.10–12 Cancer is a heterogenous disease, which embodies genetic and phenotypic variations between patients, between primary and metastatic tumors, and within a single tumor.13 We postulate that tumors secrete and/or shed a medley of tumor-derived proteins into the blood, which could be quantified, to serve as indicators of tumor burden in each patient. We previously termed these markers that are in a subset (e.g., 5-30%) of tumors as personalized tumor markers, which could each be sensitive of early signs of recurrence in a fraction of patients.14,15 The detection and measurement of proteins in blood plasma/serum is thus advantageous for mapping the unique tumor proteomic signature and portend tumor load in each patient.
Our study aims to discover and validate new personalized protein tumor markers in OvCa patients, which may be promising for detecting recurrence, to complement the utility of the conventional OvCa biomarkers CA125 and human epididymis protein 4 (HE4). In the discovery phase, we employed the proximity extension assay (PEA) technology to simultaneously profile the serum level of 1,104 proteins in HGSC patients in a 12-month follow up study. We aimed to identify a panel of personalized biomarkers (in addition to CA125 and HE4) that enable the selection of patient-tailored marker combinations for monitoring recurrence in each patient. Subsequently, we aimed to validate the ability of our top candidates, along with CA125 and HE4, to detect tumor burden changes that correlate to relapse in an independent HGSC cohort. In this study, we aimed to employ an individualized selection of the most informative markers for each patient to detect tumor burden and relapse at high sensitivity. Novel personalized biomarkers of tumor burden may aid in clinical decision-making for OvCa patients in terms of treatment duration (to eliminate residual tumor burden according to the personalized markers), treatment response, and recurrence.
All HGSC serum samples were collected and stored by the UHN Gynecologic Blood Biobank (Toronto, ON, Canada) according to a standardized protocol, to avoid systemic biases. Approval was obtained from the Research Ethics Board of UHN, Toronto, Canada (REB #10-0591).
For the discovery phase, the retrospective cohort consisted of serum samples from 30 HGSC serially collected at four timepoints (120 samples total) as follows: Timepoint 1 - Before primary debulking surgery, Timepoint 2-0.75 months after surgery, Timepoint 3 - A median of 5 months after surgery (ranged from 5 – 8 months), and Timepoint 4 - A median of 11 months (ranged from 8 – 14 months) after surgery. Twenty-five of these patients were diagnosed with recurrence at timepoint 4, according to CA125 elevation, radiological findings, or clinical symptoms. Five patients did not experience recurrence during the study period and were used as non-recurrence controls. All patients received primary debulking surgery followed by six cycles of platinum/taxane-based chemotherapy. We also included serum samples from female, age-matched, non-cancer controls (n=9), taken at three timepoints (27 samples total). This group consists of dementia patients in a 24-month follow-up study (three timepoints at month 0, month 12, and month 24), as described in a recent publication by our group (306). Although these patients were identified from an unrelated study, they were used as biological and technical references in this study for gauging how much the protein levels change between longitudinal measurements due to day-to-day biological variations and technical factors, in a non-cancer population. PEA analysis was performed for the same 1,104 analytes, in the same run, and by the same analytical provider (Olink Proteomics, Boston, MA, USA) as the HGSC study.
For the validation phase, we included retrospective, serum samples collected from 39 HGSC patients, with 4-8 timepoints (median of 6) per patient depending on availability (ranging from three months to seven years of follow-up depending on the time of relapse). Approximately 72% (28/39) of the HGSC patients received primary debulking surgery followed by six cycles of platinum/taxane-based chemotherapy, while 28% (11/39) received neoadjuvant chemotherapy with interval debulking surgery with subsequent platinum/taxane-based chemotherapy (6 cycles total). Longitudinal timepoints were chosen from those collected at pre-primary debulking surgery or pre-neoadjuvant chemotherapy, 0.75 months post-surgery or post neoadjuvant chemotherapy, and after 3rd and 6th cycle of 1st-line chemotherapy or after interval debulking and subsequent chemotherapy. Several collections every three months during remission, and collections at three months before and at imaging confirmed relapse were also included. Patients are categorized as follows: 1) Long-Term Survival/“curative” control (>7 years free of disease from last dose of chemotherapy) (n=5), 2) Platinum sensitive (relapse ~1-3 years from last dose of chemotherapy; most common HGSC case) (n=25), and 3) Platinum resistant (relapse within six months of last dose of chemotherapy) (n=9). We included patients with various timelines of recurrence to ensure our candidates correlate to relapse in a wide range of patients encountered clinically.
For the discovery phase, PEA analyses were conducted using the twelve available 92-plex panels: cardiometabolic, cell regulation, cardiovascular III, cardiovascular II, development, immune response, inflammation, metabolism, neurology, neuro-exploratory, oncology II, and organ damage. A total of 1,104 analytes were measured. The assay is described in detail in our previous publication.16
Because we are identifying personalized biomarkers in a surveillance cohort, we are interested in proteins that show significant within-individual increase upon recurrence, in each recurrent patient. Therefore, we first calculated the % rate of change in protein levels between consecutive timepoints to longitudinally assess the protein kinetics within each patient. This is calculated by the % change between timepoints divided by the number of months that elapsed. Mainly, we considered the timepoint before recurrence to the timepoint of clinically confirmed recurrence (i.e., timepoint 3-4 in the 25 recurrent HGSC patients). We calculated the protein-specific reference change value (RCV) for all 1,104 proteins based on the intra-individual biological variation (day-to-day difference) observed in the longitudinal non-cancerous controls and the analytical variation (differences between technical replicates) that was previously calculated (our unpublished data). RCV was calculated with the formula: RCV=21/2 * Z * (CV(A)2 + CV(I)2)1/2, where Z is 1.96 for 95% probability, CV(A) is the analytical variation, and CV(I) is the biological variation. RCV reflects the natural intra-individual changes across serial measurements for a given analyte. A % change in serial biomarker measurement is deemed significant if it is higher than its RCV. Our defined term of “rate of RCV” (the minimal % change per month that denotes significance) was also calculated for the purpose of this study to be comparable to the % rate of change in the proteins. We defined the rate of RCV as the RCV divided by three months since surveillance biomarkers are clinically assessed every ~three months for the first year following first-line treatment. The rate of % change in biomarker value between timepoints 3-4 (upon relapse) in the recurrent patient must exceed the “rate of RCV” of the respective protein, to be considered statistically significant. We outline below our further defined selection criteria. The candidate personalized biomarkers we selected were proteins that sequentially passed the following filtering criteria that we pre-defined and were determined a priori (proteins remaining means proteins that meet the predefined criteria).
1. More than 100% decrease from timepoint 1 (pre-surgery) to timepoint 2 (0.75 months post-surgery) or timepoint 3 (5 months post-surgery; post-chemotherapy), in at least one patient (642/1,104 proteins remaining). This selects for proteins that significantly decreased due to a decrease in tumor burden, following primary debulking surgery and/or chemotherapy.
2. % Rate of change from timepoint 3-4 is greater than the rate of RCV in at least one recurrent patient (306/1,104 proteins remaining). This ensures the within-individual change upon relapse is statistically significant, after accounting for biological and analytical variations.
3. % Rate of change from timepoint 3-4 is less than the rate of RCV in all non-recurrent patients (263/1,104 proteins remaining). This removed 43 proteins that showed non-specific increase from timepoint 3-4 in any non-recurrent patient.
4. More than 100% increase between timepoint 3-4 in at least one recurrent patient (201/1,104 proteins remaining). This stringent criterion selects for proteins with more than two-fold increase upon relapse.
5. More than 70 Normalized Protein Expression value (NPX is a relative unit of amount of protein expression for PEA analysis) at timepoint 4 (i.e. relapse) in at least one recurrent patient (103/1,104 proteins remaining). In our previous study on the technical performance of PEA, we observed a higher analytical variability for protein measurements with low NPX values (<2 times the lower limit of detection (LLD)]. Therefore, we chose this arbitrary cut-off to select for proteins that showed expression at least two-times above the LLD (LLD of the 1,104 analytes ranged from 3.7 – 34.4 NPX). This ensures the protein expression at relapse and % change observed from timepoint 3-4 are analytically reliable.
6. We narrowed down the comprehensive list of 103 candidates to the top candidates most suitable for further validation. To achieve this, we selected proteins that increased upon relapse in most of the recurrent patients. We also identified proteins that increased by the highest % of NPX upon relapse, per recurrent patient. Employing the principle of parsimony, among the 103 candidates, we selected the least number of proteins that together were informative for relapse by the highest % of patients (the goal was achieving at least 90%). This resulted in a final list of 23 top candidate personalized markers, in addition to the known markers CA125 and HE4, which also passed our filtering criteria.
PEA analyses were conducted using the seven 92-plex panels. A total of 644 proteins were measured. Unfortunately, we could only perform PEA analysis for seven 92-plex panels due to cost constraints. We selected the panels that covered most of our 23 candidates, which included 21 candidates. Unfortunately, two candidates, DNAJB1 and PSIP1 were not included in the seven panels and were not analyzed further in our validation study.
We were interested in assessing if the 21 out of the 23 candidate proteins that we identified in our discovery phase, in addition to CA125 and HE4, showed significant within-individual increase upon recurrence in the 35 recurrent HGSC patients in an independent validation study. As per the discovery study, we first calculated the % rate of change in the 21 proteins between sequential timepoints, to longitudinally assess the protein kinetics within each patient. Mainly, we considered the timepoint immediately before recurrence to the timepoint of clinically confirmed recurrence for each patient. We used the protein specific RCV and rate of RCV that was previously calculated in the discovery study. The % rate of change in the 21 proteins from timepoint before relapse to relapse in the recurrent patient must exceed the rate of RCV of the respective protein to be considered statistically significant.
To select a refined panel of personalized markers, we performed the same analysis as described for the 21 candidate proteins, applied to all 644 proteins measured in the validation study. In addition, we outline below our further pre-defined selection criteria. The candidate personalized biomarkers we selected were proteins that sequentially passed the following filtering criteria:
1. % Rate of change from the timepoint immediately before relapse to relapse is greater than the rate of RCV in at least one recurrent patient (466/644 proteins remaining).
2. % Rate of change from before last to the last timepoint is less than the rate of RCV in all non-recurrent patients (450/644 proteins remaining). This removed 16 proteins that showed non-specific increase in any non-recurrent patient.
3. Decreased from the first timepoint (baseline/pre-surgery/pre-chemotherapy) to timepoint after six cycles of chemotherapy in at least one recurrent patient (391/644 proteins remaining).
4. More than 50% increase between the timepoint before relapse to relapse in at least one recurrent patient (256/644 proteins remaining).
5. The NPX at relapse is greater than two times the LLD of the corresponding protein in at least one recurrent patient (231/644 proteins remaining).
6. We narrowed down the comprehensive list of 231 candidates to the top candidates for a refined panel of personalized tumor markers. To achieve this, we included the 18 candidates that we previously identified in the discovery phase, which also were verified in this validation study. We also identified additional proteins that increased by the highest % upon relapse per recurrent patient. We again selected the least number of proteins that together were informative for the highest % of patients (achieving at least 90%). This resulted in a final list of 33 top candidate personalized markers, in addition to the known markers CA125 and HE4.
Statistical analyses were performed using the GraphPad Prism software version 9.0.2 (GraphPad Software, San Diego, CA) and R statistical software version 4.0.4 (www.rproject.org). The Kruskal-Wallis test with Bonferroni’s multiple correction was employed to compare the % rate of change in protein level from timepoint 3 to 4 between three groups: 1) Elevated CA125 at recurrence, 2) Non-elevated CA125 at recurrence, and 3) Non-recurrence. Pearson correlation analysis was performed to examine the relationship between % rate of change in protein level from timepoint 3 to 4 and age at diagnosis. The Kruskal-Wallis test was used to assess the relationship between the % rate of change in protein level from timepoint 3 to 4 between clinical groups (BRCA1/2 mutation status and residual tumor volume). P-values of less than 0.05 were considered to be statistically significant.
Demographics and clinicopathological characteristics of the 30 HGSC patients in the discovery study cohort are summarized in Table 1. For one patient (LR45) the CA125 value was not elevated (<35 U/mL) at diagnosis and thus CA125 monitoring would not be recommended for surveillance. In our study, 17/25 (68%) of recurrent patients showed increase in CA125 above the reference range of 35 U/mL, which strongly indicates biochemical relapse, and would prompt referral to imaging in standard clinical practice. It is important to note that a median of 6 months elapsed between timepoint 3-4 (ranges from 3-9 months). Therefore, it is unclear whether the CA125 value doubled in the other patients in the clinical follow-up prior to clinical relapse (3 months period). We calculated the % change in 3 months in clinical CA125 value based on the change from T3-4 and the number of elapsed months. Patients LR22 and LR45 did not show a % increase by ≥100% (CA125 did not double) in 3 months. Patients LR38, LR39, LR42, and LR43 did show % increase by >100% (at least doubling in CA125 value) in 3 months.
| Clinically assessed serum CA125 concentration (U/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient ID | Age at diagnosis | Germline BRCA1/2 mutation | Residual tumor volume after primary surgery | T 1 (0 months, pre-surgery) | T 2 (0.75 months post-surgery) | T 3 (~5 months post-surgery) | T 4 (~11 months post-surgery) | % change in 3 months (based on T 3-4) | Clinical recurrence at T 4?** |
| HR14 | 61 | – | None | 178 | 86 | 13 | 40 | 69 | Y |
| HR20 | 81 | – | None | 448 | 121 | 27 | 67 | 148 | Y |
| HR21 | 67 | Not noted | >1 cm | 328 | 154 | 49 | 328 | 569 | Y |
| HR23 | 68 | – | None | Not noted | 661 | 9 | 1168 | 6439 | Y |
| HR24 | 47 | + | Not noted | 15276 | 2051 | 67 | 113 | 34 | Y |
| HR25 | 67 | – | >1 cm | 436 | 154 | 8 | 132 | 1550 | Y |
| HR26 | 61 | – | None | 1454 | 410 | 13 | 145 | 338 | Y |
| HR27 | 69 | + | >1 cm | 362 | 235 | 27 | 371 | 956 | Y |
| HR28 | 68 | – | Not noted | 144 | 148 | 16 | 72 | 150 | Y |
| HR30 | 65 | – | None | 92 | 81 | 11 | 44 | 150 | Y |
| HR31 | 54 | – | None | 92 | 62 | 45 | 139 | 157 | Y |
| HR34 | 56 | + | <1 cm | 473 | 36 | 5 | 51 | 460 | Y |
| HR35 | 64 | Not noted | Not noted | 346 | 40 | 12 | 93 | 225 | Y |
| HR40 | 46 | – | >1 cm | 8453 | 4057 | 72 | 165 | 65 | Y |
| HR46 | 72 | – | <1 cm | 423 | 260 | 63 | 81 | 29 | Y |
| HR5 | 40 | – | None | 2659 | 287 | 29 | 57 | 32 | Y |
| HR8 | 42 | + | None | 2142 | 233 | 6 | 488 | 4017 | Y |
| LR22 | 66 | – | >1 cm | 129 | 85 | 11 | 29 | 55 | Y |
| LR36 | 49 | + | >1 cm | 1371 | 1164 | 6 | 8 | 17 | Y |
| LR37 | 46 | – | None | 423 | 70 | 22 | 30 | 36 | Y |
| LR38 | 67 | + | None | 233 | 7 | 2 | 18 | 400 | Y |
| LR39 | 51 | – | None | 7628 | 167 | 11 | 34 | 105 | Y |
| LR42 | 68 | – | None | 362 | 25 | 3 | 14 | 122 | Y |
| LR43 | 42 | – | None | 935 | 77 | 5 | 25 | 240 | Y |
| LR45* | 63 | – | Not noted | 9 | 7 | 6 | 12 | 43 | Y |
| NR10 | 61 | + | None | 1060 | 119 | 8 | 6 | -13 | N |
| NR2 | 57 | – | None | 3019 | 96 | 9 | 16 | 26 | N |
| NR4 | 39 | – | >1 cm | 2833 | 339 | 3 | 10 | 78 | N |
| NR6 | 64 | – | Not noted | 2121 | 32 | 5 | 5 | 0 | N |
| NR9 | 73 | – | <1 cm | 1162 | 75 | 7 | 5 | -14 | N |
Demographics and clinicopathological characteristics of the 39 HGSC patients in the validation cohort are summarized in Table 2. CA125 was non-elevated at diagnosis in 13% (5/39) of the HGSC cases, representing patients who would not be recommended for CA125 monitoring by US and international guidelines and have no conventional biomarkers for surveillance.12
| Clinically assessed serum CA125 concentration (U/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient ID | Age at diagnosis | Germline BRCA1/2 mutation | Residual tumor volume after primary surgery | Baseline (at diagnosis) | Post-chemotherapy (6 cycles total) | Before relapse (or before last timepoint in non-recurrence) | At relapse** (or last timepoint in non-recurrence) | % change in 3 months |
| Platinum sensitive (n=25) | ||||||||
| PS1 | 60 | – | >1 cm | 1350 | 6 | 14 | 15 | 7 |
| PS11 | 58 | + | <1 cm | 139 | 9 | 20 | 22 | 10 |
| PS12* | 46 | – | Not noted | 13 | 5 | 3 | 35 | 1067 |
| PS15 | 42 | + | None | 212 | 5 | 11 | 26 | 136 |
| PS17 | 50 | – | None | 731 | 8 | 6 | 83 | 1283 |
| PS19 | 52 | – | None | 938 | 3 | 28 | 72 | 157 |
| PS2 | 66 | – | <1 cm | 1175 | 13 | 11 | 33 | 200 |
| PS21 | 56 | + | None | Not noted | 6 | 6 | 16 | 167 |
| PS22 | 53 | Not noted | None | 6458 | 6 | 12 | 90 | 650 |
| PS23 | 55 | Not noted | Not noted | 434 | 2 | 3 | 28 | 833 |
| PS24 | 65 | – | Not noted | Not noted | 23 | 6 | 32 | 433 |
| PS26 | 61 | – | >1 cm | 1454 | 13 | 22 | 145 | 559 |
| PS27 | 54 | – | None | 2229 | 8 | 12 | 39 | 225 |
| PS29 | 47 | + | None | 835 | 10 | 14 | 22 | 57 |
| PS31 | 39 | – | >1 cm | 2833 | 3 | 10 | 51 | 410 |
| PS32* | 64 | + | <1 cm | 23 | 5 | 24 | 63 | 163 |
| PS34 | 64 | Not noted | None | 1060 | 8 | 11 | 25 | 127 |
| PS35 | 58 | Not noted | None | 424 | 4 | 3 | 56 | 1767 |
| PS36* | 67 | Not noted | None | 14 | 9 | 21 | 30 | 43 |
| PS38 | 63 | + | None | 466 | 12 | 10 | 74 | 640 |
| PS39 | 52 | + | None | Not noted | 5 | 97 | 199 | 105 |
| PS41 | 68 | – | None | Not noted | 11 | 11 | 1168 | 10518 |
| PS42 | 65 | – | None | 64 | 4 | 12 | 70 | 483 |
| PS7 | 68 | – | None | 248 | 12 | 12 | 76 | 533 |
| PS9* | 65 | – | Not noted | 11 | 17 | 12 | 22 | 83 |
| Platinum resistant (n=9) | ||||||||
| PR1 | 74 | – | None | 1004 | 14 | 11 | 14 | 27 |
| PR11 | 77 | Not noted | <1 cm | 167 | 10 | 10 | 40 | 300 |
| PR12 | 57 | + | None | 1975 | 32 | 22 | 1126 | 5018 |
| PR14 | 59 | – | None | Not noted | 7 | 13 | 64 | 392 |
| PR29 | 49 | + | >1 cm | 1371 | 38 | 6 | 18 | 200 |
| PR32 | 64 | – | >1 cm | 362 | 235 | 30 | 371 | 1137 |
| PR4 | 66 | – | <1 cm | 1187 | 20 | 22 | 60 | 173 |
| PR7 | 63 | Not noted | <1 cm | 3351 | 8 | 8 | 20 | 150 |
| PR8 | 81 | – | None | 448 | 32 | 32 | 67 | 109 |
| Long term survival (n=5) | ||||||||
| LTS17 | 67 | – | None | 159 | 8 | 9 | 8 | -11 |
| LTS18 | 74 | – | None | Not noted | 8 | 9 | 21 | 133 |
| LTS20 | 50 | – | None | Not noted | 8 | 4 | 11 | 175 |
| LTS21* | 53 | – | None | 9 | 5 | 5 | 5 | 0 |
| LTS24 | 55 | – | None | 383 | 9 | 7 | 9 | 29 |
We employed a patient-centric analysis to identify the top proteins that sensitively reflect changes in tumor burden upon relapse in individual patients. We identified a panel of the top 23 candidate personalized markers, in addition to CA125 and HE4. The names and biological functions of the 23 candidate proteins are listed in Table 3. The 23 candidate personalized markers each showed a sensitivity of 16-52% at 100% specificity for detecting relapse (Figure 1). The % change in 3 months (for clinical relevance, as biomarkers are usually assessed every ~3 months) for each personalized marker and patient are shown in Figure 1. The marker is considered to signify significant risk of relapse (shown in orange) for the patient, if it showed a % change greater than the RCV and the maximum % change seen in all non-recurrent patients was less than the RCV (100% specificity). In addition to this criterion, markers that showed a % change greater than two-times the RCV, are considered to denote a highly significant risk of relapse in that patient (shown in red).
| Protein name (Abbrev.) | Protein name (full) | Location* | Function** |
|---|---|---|---|
| CCL20 | C-C motif chemokine 20 | Secreted to blood | Serves as a ligand for C-C chemokine receptor 6 (CCR6), where the ligand-receptor pair is responsible for the chemotaxis of T-cells and B- cells and plays a vital role in inflammation. |
| CHI3L1 | Chitinase-3-like protein 1 | Intracellular, Locally secreted (different isoforms) | May have a role in inflammation and apoptosis |
| DEFB4A | Defensin, beta 4A | Locally secreted | Anti-microbial activity. May act as a ligand for CCR6. |
| DNAJB1 | DnaJ homolog subfamily B member 1 | Intracellular | Regulates heat-shock proteins. |
| EGF | Pro-epidermal growth factor | Membrane | Stimulates the growth of various epidermal and epithelial tissues. |
| EIF4EBP1 | Eukaryotic translation initiation factor 4E-binding protein 1 | Intracellular | Mediates protein translation of hormones, growth factors etc. in the MAP kinase and mTORC1 pathways. |
| FKBP5 | Peptidyl-prolyl cis-trans isomerase FKBP5 | Intracellular | Involved in the intracellular trafficking of steroid hormone receptors. |
| KLK11 | Kallikrein-11 | Intracellular, Locally secreted (different isoforms) | Multifunctional protease. |
| KLK6 | Kallikrein-6 | Intracellular, Locally secreted (different isoforms) | Multifunctional serine protease. |
| MSLN | Mesothelin | Intracellular, Membrane (different isoforms) | Membrane forms may be involved in cell adhesion. |
| PDGFA | Platelet-derived growth factor subunit A | Intracellular, Secreted to blood (different isoforms) | Plays important role in controlling cell proliferation, migration, survival, and chemotaxis. |
| PMVK | Phosphomevalonate kinase | Intracellular | Enzyme that catalyzes step in isoprenoid biosynthesis. |
| PSIP1 | PC4 and SFRS1-interacting protein | Intracellular | Transcriptional coactivator involved in neural stem cell differentiation and neurogenesis. |
| PVRL4 | Nectin-4 | Membrane, Locally secreted (different isoforms) | Possibly involved in cell adhesion. |
| SDC1 | Syndecan-1 | Intracellular, Membrane, Secreted to blood (different isoforms) | Not well studied. Cell surface proteoglycan that regulates exosome biogenesis. |
| SIRT2 | NAD-dependent protein deacetylase sirtuin-2 | Intracellular | Regulates diverse biological processes, such as microtubule dynamics and cell differentiation. Major role in controlling cell cycle progression and genomic stability. |
| STK4 | Serine/threonine-protein kinase 4 | Intracellular | Well-studied pro-apoptotic kinase that is a vital component of the Hippo signaling pathway, which is critical for tumor suppression, restricting proliferation, and encouraging apoptosis. |
| TBCB | Tubulin-folding cofactor B | Intracellular | Involved in alpha-tubulin folding and tubulin synthesis. |
| TCL1A | T-cell leukemia/lymphoma protein 1A | Intracellular | Promotes cell proliferation and survival. |
| TFPI2 | Tissue factor pathway inhibitor 2 | Intracellular, Secreted to extracellular matrix (different isoforms) | Possibly involved in regulating matrix remodeling. |
| TGM2 | Protein-glutamine gamma-glutamyltransferase 2 | Intracellular | Involved in protein cross-linking. |
| TNFSF14 | Tumor necrosis factor ligand superfamily member 14 | Intracellular, Membrane, Secreted to blood (different isoforms) | Cytokine that stimulates T cell proliferation. |
| TOP2B | DNA topoisomerase 2-beta | Intracellular | Regulates DNA topological states and forms double-strand breaks. |
* Predicted protein location according to Human Protein Atlas (https://www.proteinatlas.org/).
** Putative protein function based on STRING (https://string-db.org/).
To complement CA125, the panel of 23 personalized markers, plus HE4 (24 proteins in total), was used to select a custom marker combination for predicting relapse in each patient (combinations comprised of unique 1-19 informative markers per patient). These patient-centric marker combinations predicted relapse in 92% of patients. Nine of the 25 recurrent patients had at least one personalized marker (ranging from 1-11 markers) that showed larger % increase upon relapse compared to CA125, as measured by PEA.
PSIP1 and STK4 showed weak but significant correlation between % change upon relapse and age at diagnosis (r = 0.57 and 0.49, respectively). None of the 23 candidate personalized markers showed significant association between % change upon relapse with BRCA1/2 mutation status and residual tumor volume after primary or interval debulking surgery.
We examined the sensitivity of the 21 candidate proteins in this independent HGSC cohort. In Figure 2 we show a heatmap of their % change in 3 months per patient.
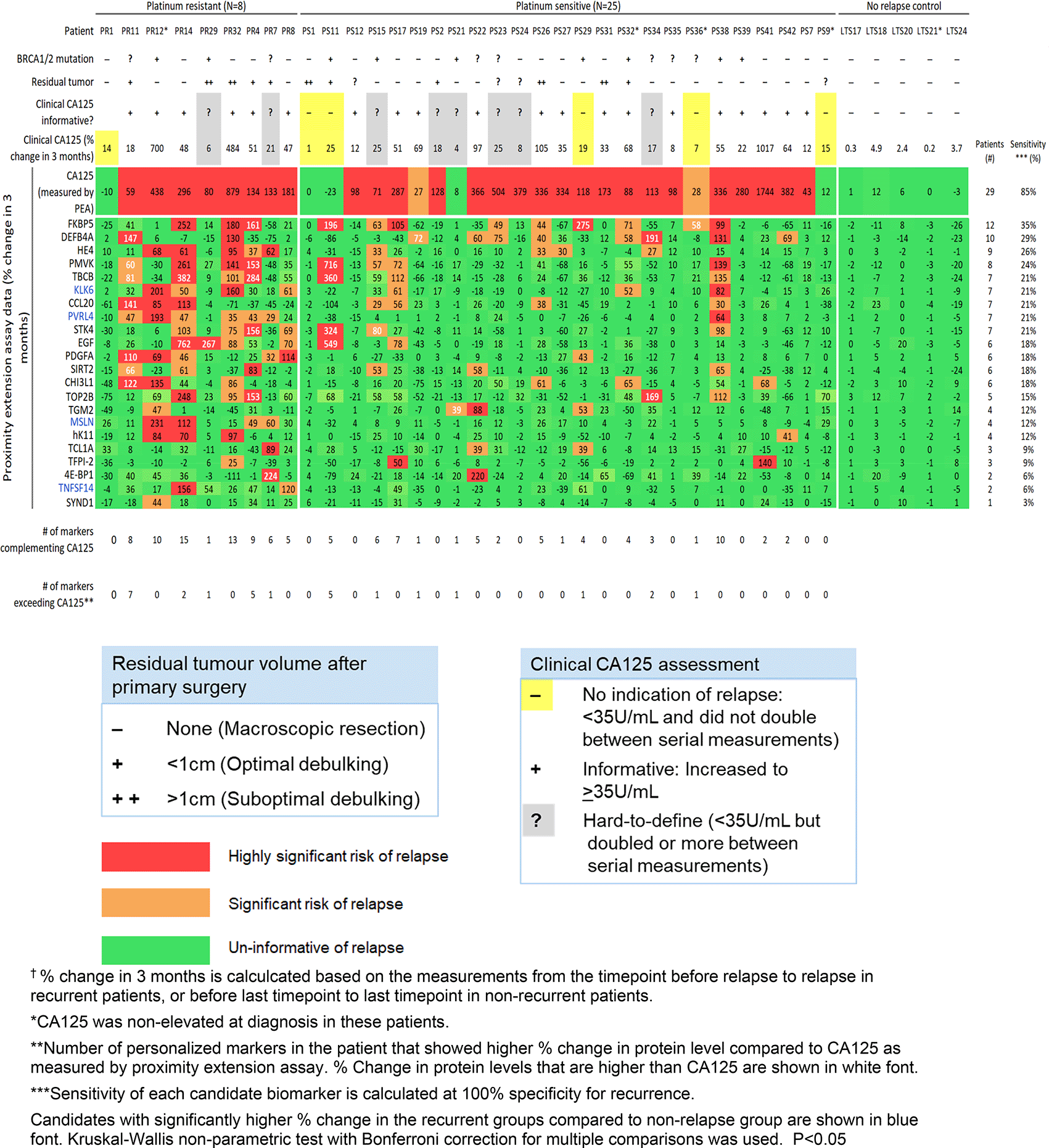
The 21 candidates we previously identified in the discovery study each showed a sensitivity of 3-35% (at 100% specificity) for detecting relapse. To complement CA125, our previously identified panel of the 21 candidate personalized markers, plus HE4, was used to select a custom marker combination for predicting relapse in each patient (combinations comprised of unique 1-13 informative markers per patient). These patient-centric marker combinations predicted relapse in 71% (24/34) of the recurrent patients. Twelve of the 34 recurrent patients had at least one personalized marker (ranging from 1-7 markers) that showed larger % increase upon relapse compared to CA125 as measured by PEA.
Using patient-centric selection criteria, we identified a panel of the top 33 candidate personalized markers, in addition to CA125 and HE4. This panel includes 18 candidates that were identified in our discovery cohort. The names and biological functions of the 15 additional candidate proteins are listed in Table 4. The 33 candidate personalized markers each showed a sensitivity of 6-41% at 100% specificity for detecting relapse (Figure 3). To complement CA125, the panel of 33 personalized markers, plus HE4 (34 proteins total), was used to select a custom marker combination for predicting relapse in each patient (combinations comprised of unique 1-22 informative markers per patient). These patient-centric marker combinations predicted relapse in 91% of patients. Eighteen of the 34 recurrent patients had at least one personalized marker (ranging from 1-11 markers) that showed larger % increase upon relapse compared to CA125 as measured by PEA.
| Protein name (Abbrev.) | Protein name (full) | Location* | Function** |
|---|---|---|---|
| AGR2 | Anterior gradient protein 2 homolog | Intracellular | Normally promotes cell adhesion but is a known proto-oncogene that may be involved in cell differentiation, proliferation, and migration. |
| ANGPTL4 | Angiopoietin-related protein 4 | Intracellular, Secreted to blood | Play a role in regulating angiogenesis and tumorigenesis, such as reducing proliferation and vascular leakage. |
| DECR1 | 2,4-dienoyl-CoA reductase, mitochondrial | Intracellular | Enzyme for beta-oxidation and the metabolism of unsaturated fatty enoyl-CoA esters. |
| DSG4 | Desmoglein-4 | Membrane | Part of intercellular desmosome junctions and is involved in mediating cell-cell adhesion. |
| EREG | Proepiregulin | Membrane, Secreted to blood | Ligand of the EGF receptor. Promotes cell growth, proliferation, angiogenesis, and inflammation. |
| IL4 | Interleukin-4 | Secreted to blood | Plays a role in the activation processes of many B-cell and other cell types. |
| MMP9 | Matrix metalloproteinase-9 | Secreted to blood | May function in proteolysis in the extracellular matrix and in leukocyte migration. |
| NCF2 | Neutrophil cytosol factor 2 | Intracellular | The cytosolic component of the multi-protein complex, NADPH oxidase, found in neutrophils |
| NPPB | Natriuretic peptides B | Secreted to blood | Cardiac hormone that may function in cardiovascular homeostasis. |
| OSM | Oncostatin-M | Intracellular, Secreted to blood | Regulates cell growth and cytokine production. |
| PTX3 | Pentraxin-related protein PTX3 | Secreted to blood | Involved in innate immunity and inflammatory responses and possibly plays a role in the female reproductive system. |
| PVALB | Parvalbumin alpha | Intracellular | Functions in muscle relaxation after contraction. |
| SERPINA12 | Serpin A12 | Locally secreted | Adipokine which regulates insulin activity. |
| SULT1A1 | Sulfotransferase 1A1 | Intracellular | Sulfotransferase enzyme which catalyzes many hormones, neurotransmitter, and drugs. Plays a role in the activation of carcinogenic N-hydroxyarylamines to DNA binding products and thereby modulating cancer risk |
| VIM | Vimentin | Intracellular | Intermediate filaments located in various non-epithelial cells, especially mesenchymal cells. |
* Predicted protein location according to Human Protein Atlas (https://www.proteinatlas.org/).
** Putative protein function based on STRING (https://string-db.org/).
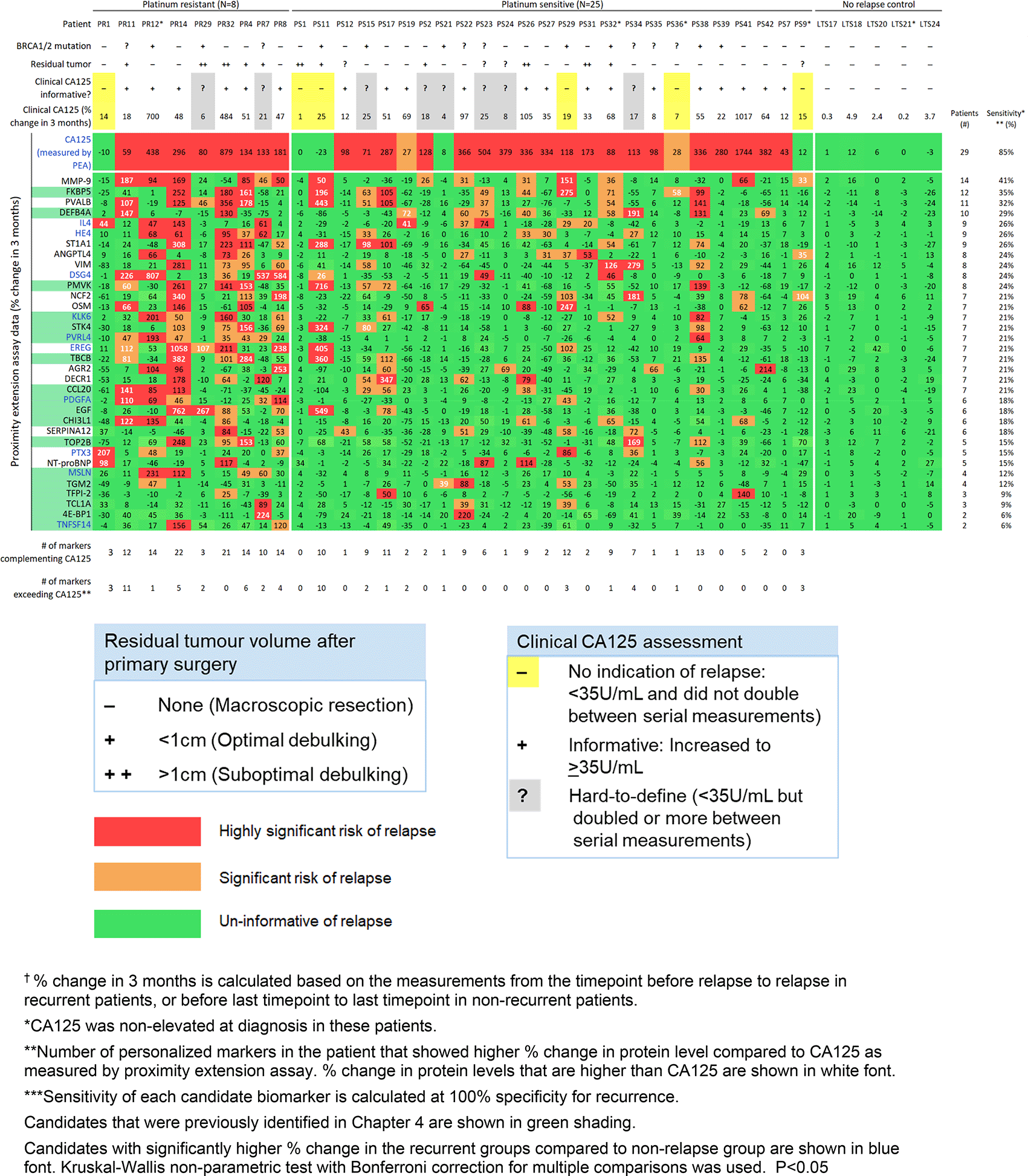
Three of the 33 candidate personalized markers (TBCB, PMVK, FKBP5) showed significant association between the % change upon relapse and BRCA1/2 mutation status. KLK6 and PTX3 showed significant but weak correlation of % change upon relapse with age at diagnosis (r = 0.34 and 0.35, respectively). The % change upon relapse was not associated with residual tumor volume for any of the markers.
Building on our emergent concept of personalized tumor markers,14,15 we conducted an exploratory discovery study and a preliminary follow-up validation study to identify potential personalized markers that may correlate to tumor burden and predict relapse in HGSC. For the exploratory phase, conducting a thorough literature search revealed that all 23 identified proteins, except DEFB4A, PMVK, and TBCB, have been widely implicated in OvCa at either the genome, transcriptome, or proteome level. Nevertheless, DEFB4A has been reported as a candidate prognostic biomarker for colorectal cancer and have shown polymorphisms in colorectal cancer patients, which suggests a role in inter-individual cancer heterogeneity.17,18 A small mass spectrometry-based proteomics study found that an increased level of PMVK in chemotherapy-naïve HGSC tissue was associated with favorable prognosis (progression-free survival of ≥ 18 months; n=6 samples).19 TBCB is an essential protein for microtubule folding and cellular stability and is phosphorylated by spleen tyrosine kinase (SYK) in paclitaxel-resistant ovarian tumor cells.20
There have been 10 studies employing PEA technology to screen for over 92 proteins in OvCa serum or plasma to identify key players and biomarkers.21–30 Ours is the first reported study to use all 12 available PEA panels for profiling 1,104 proteins in OvCa. Moreover, the focus so far has been on identifying diagnostic biomarkers of early detection for OvCa.22,23–25,27–30 Our study takes a different angle of identifying personalized markers that are highly sensitive for detecting tumor burden but in the form of recurrent, instead of primary disease. Amongst our top 23 candidate proteins, KLK11, TFPI2, TNFSF14, KLK6, PVRL4, and PDGFA were selected as potential diagnostic biomarkers of OvCa in previous studies employing PEA analysis.21–23,25 KLK11, TNFSF14, and PVRL4 were significantly elevated in the serum or plasma of early-stage OvCa in multiple PEA studies, which validates their likely role in ovarian tumorigenesis and their elevation in the circulation at an early stage.21–23,25
Some of our 23 candidate proteins are well-studied in OvCa. Cisplatin stimulation during first-line chemotherapy has been shown to trigger an increase in CCL20 production by ovarian tumor-associated macrophages.31–35 CCL20 activates C-C chemokine receptor 6 (CCR6) on ovarian tumor cells and, in turn, promotes epithelial-to-mesenchymal transition and cancer cell migration.31–35 High expression of CHI3L1 is associated with poor prognosis and chemoresistance in OvCa.36–39 CHI3L1 induces AKT and ERK signaling pathways to promote OvCa stem-like cells.36–39 TGM2 is a player in the chromatin-binding protein family, high-mobility-group-box (HMGB), interactome in OvCa. TGM2 is correlated to poor outcome and promotes epithelial-to-mesenchymal transition.40–43 STK4 plays a key role in the kinase cascade elicited in the Hippo signaling pathway, which controls organ growth.44–47 Deregulation of the Hippo signaling pathway is implicated in ovarian carcinogenesis.44–47 SDC1 serves as a receptor for collagen alpha-1(I) chain (COL1A1) proteins secreted by fibroblasts. Ligand-receptor binding of SDC1 and COL1A1 activates AKT signaling pathways to promote OvCa cell motility.48–50
Our goal was to evaluate the ability of the top 21 of the 23 personalized marker candidates to detect relapse in a larger, independent, longitudinal HGSC cohort. The 21 candidates demonstrated a significant increase in biomarker value upon relapse in at least 1 recurrent HGS patient in the validation cohort (Figure 2). However, the 21 candidates showed lower sensitivity (3-35%, at 100% specificity for recurrence) for detecting relapse in the validation cohort compared to the discovery findings (16–52%). Personalized marker combinations detected relapse in 71% of patients in the validation cohort, compared to 92% in the discovery study. The discrepancy between the two findings is likely due to the limited sample sizes. As personalized markers are defined by being expressed in a small subset (5-30%) of tumors and are not informative for most patients of a cancer type, a large sample size is needed to accurately encompass the heterogeneity that exists in ovarian cancer. Our discovery and validation studies provide the necessary data for calculating an accurate sample size that would provide statistical power in future large-scale prospective studies. Studies have shown differential molecular profiles for platinum resistant (relapse in <6 months) and platinum sensitive (relapsed in > 6 months) cases.51,52 The validation data may better reflect the biomarkers’ performance for surveillance in clinical settings, where there is currently no clinically used predictor of platinum sensitivity/resistance.53 Finally, the validation cohort may represent a different subset of tumor composition, response to chemotherapy, and molecular drivers for relapse.54
Our patient-centric analysis selected 18 of the top 21 candidates that we identified in our discovery phase, which we included in our refined panel of the top 33 personalized tumor markers (Figure 3). The 15 new candidate personalized markers each showed a sensitivity of 15-41% at 100% specificity for tracking recurrence. Further validation studies are needed for these 15 candidates. The early detection of recurrence, even before recurrent tumors can be detected through imaging modalities, may provide benefit on survival outcome, as there are increasing selections of maintenance treatment and experimental targeted trials for patients to enroll in.8,55–58
We recognize that our study has important limitations, as follows:
1. The sample sizes of both the discovery and validation cohorts are relatively small (<40 patients). One reason is that each patient has contributed multiple longitudinal samples and the expense of running all of the PEA panels is high, at around $1,000 USD per sample. Fortunately, once a panel is defined, consisting of <40 candidate markers, the cost is more realistic (estimated at approx. $100 USD per sample) for large-scale clinical validation and clinical use.
2. It has not yet been shown that intense surveillance post-primary treatment extends patient survival or quality of life.1,2
Studies have shown that initiating aggressive second-line therapy at time of elevated CA125, before imaging is suggestive of relapse, does not improve outcome.59 Our goal is to personalize ovarian cancer surveillance to determine the optimal decision points for prompting referral to imaging and consequently initiating second-line treatment in each patient.
Our discovery phase identified a panel of 23 candidate personalized tumor markers that generated patient-centric marker combinations (comprised of 1-19 informative markers per patient) for detecting recurrence with 92% sensitivity. In our preliminary validation study, we identified 33 candidate personalized markers that each was informative of relapse in 6-41% of recurrent patients. Our study validated the ability to detect relapse for 18 identified candidates. Among the 33-personalized marker panel (plus HE4), we identified unique, custom marker combinations (1-22 markers per patient) that were informative of recurrent disease in 91.2% of patients.
In the future, personalized tumor protein profiles generated by multiplexed proteomics technologies could be monitored for changes in expression that may trigger clinically actionable events, such as selecting the best treatment, calculating prognostic risk, and detecting recurrence in the patient.60
Harvard Dataverse. Discovery and preliminary validation of a new panel of personalized ovarian cancer biomarkers for individualized detection of recurrence. doi: 10.7910/DVN/TSLJBW. 61
This project contains the following underlying data:
• Discovery dataset_proximity extension assay_data in NPX_Serum.xlsx (Raw data from the proximity extension assay for the discovery study).
• Validation dataset_proximity extension assay_data in NPX_Serum.xlsx (Raw data from the proximity extension assay for the validation study).
Data is available under the terms of the CC0 1.0 Deed (CC0 1.0 Universal).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Breast and ovarian cancer, circulating markers, genomics
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: High-grade serous ovarian cancer, bioinformatics, genetics
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Ovarian cancer biomarkers
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 23 Nov 23 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)