Keywords
Graphical User Interface (GUI), machine learning, radiomics, pyradiomics, plastimatch, 3DSlicer
Graphical User Interface (GUI), machine learning, radiomics, pyradiomics, plastimatch, 3DSlicer
As per the World health organization (WHO), cancer is the second leading cause of death worldwide. Globally one out of six deaths are caused by cancer alone which amounts to 9.6 million deaths in 2018 (WHO-Cancer, 2022; Ferlay et al., 2019). Treatment of cancer has always remained a challenging task for the oncology community. Although earlier diagnosis and treatment are often associated with a better outcome of treatment, at the same time selection of appropriate patients for appropriate treatment is important (Stewart et al., 2018). Considering the complexity of this disease and treatment, oncology is gradually moving towards personalized medicine (Agyeman and Ofori-Asenso, 2015). A pathological test is considered a confirmatory test for cancer. Imaging tests like Computed Tomography (CT)/Positron Emission Tomography (PET)/Magnetic Resonance Imaging (MRI) also play an important role in diagnosis, treatment planning, and follow-up of the disease (Caers et al., 2014). In the last several years, the role of imaging and various advanced tests like immunohistochemistry (IHC), polymerase chain reaction (PCR), and gene sequencing has been increasing gradually in personalizing the treatment. Similarly, the role of artificial intelligence (AI) and radiomics has also witnessed a surge in oncology research over the last few years. Radiomics has been identified as an area of research to develop imaging biomarkers for the personalized treatment of cancer (Lambin et al., 2012; Aerts et al., 2014; Goodwin et al., 2017). Radiomics is a method to extract high throughput data from medical images. These features have the potential to uncover disease characteristics that are not appreciated by the expert radiologist or imaging personnel through visual interpretation (Yip et al., 2017). As radiomic features are extracted directly from medical images it provides a non-invasive method for tumor characterization as demonstrated by various researchers in the past (Lambin et al., 2012; Goodwin et al., 2017; Aerts et al., 2014). Researchers have shown the role of radiomics as a clinical predictor helping in advanced cancer care as personalized medicine in cancer (Haider et al., 2020). Apart from its role in precision diagnoses and characterization of a tumor, the role of radiomics has also been demonstrated in treatment planning (Limkin et al., 2017).
Several open-source and licensed software packages for radiomic extraction, like IBEX, RaCaT, CaPTK, LifeX or CGITA, Pyradiomics, and TexRad have been developed and used by several researchers in the past (Pfaehler et al.,2019; Nioche et al., 2018; Zhang et al., 2015; Davatzikos et al., 2018; Johnson, 2015; Haralick et al., 1973).
The main challenges with these software packages are the complexity of radiomic extraction and the lack of standardized feature extraction from this software. The mathematical feature definitions have been provided image biomarker standardization initiative (IBSI) to standardize the radiomic extraction. It has also provided the phantom data sets with radiomic feature values (Zwanenburg, 2017; Zwanenburg et al., 2016). Although IBSI standard is widely known, only a very few radiomic softwares have standardized the entire radiomic pipeline for feature extraction. Furthermore, the majority of these radiomic extractors have operability issues and not all defined features are extracted as defined by IBSI. Pyradiomics is a widely-used open source radiomics package and adheres to the IBSI standards, but it is not user-friendly. For instance, Pyradiomics is run on a command prompt and customization is technically demanding. Hence, the use of this software is cumbersome and technically demanding for clinical doctors or scientists.
In this study, we have developed and validated a graphical user interface (GUI) based radiomic extraction software using the pyradiomics package. This software adheres to all features defined by the IBSI.
This work is part of the Big Imaging data approach for Oncology in a Netherlands India Collaboration (BIONIC) and “personal health Train for radiation oncology in India and the Netherlands” (TRAIN) project is approved by the IEC of the hospital as a retrospective study. This software (PyRadGUI) was developed using Python open-source software on Windows systems.
This study was approved by the hospital Institutional Ethics Committee (Institutional Ethics Committee-I, Tata Memorial Centre [IEC, TMC], Mumbai, India Approval Number: 1905; dated: October 05, 2017) as a retrospective study, with waivers of informed consent from involved patients as per IEC policy of our hospital by the same Ethics Committee.
PyRadGUI front end was developed by using the open-source software Python 3.6.5 (Python, 2022) and the Python tk8.6 (Python tk, 2022) module was used for the development of the graphical user interface (GUI). The open source Plastimatch package 1.8.0 (Plastimatch, 2022) was used for Digital Imaging and Communications in Medicine (DICOM) to Nearly Raw Raster Data (NRRD) conversion of imaging data and the radiomic package in python; Pyradiomics 3.0 (Griethuysen et al., 2017) was used for radiomic feature extraction. This software loads DICOM images and region of interest (ROI) from a specific folder on the computer. First, it converts the image and ROI in NRRD format using Plastimatch 1.8.0, subsequently, it extracts radiomic features from the image and finally stores the output in comma separated values (CSV) format in an output folder on the computer. The details of the software are described below (Figure 1).
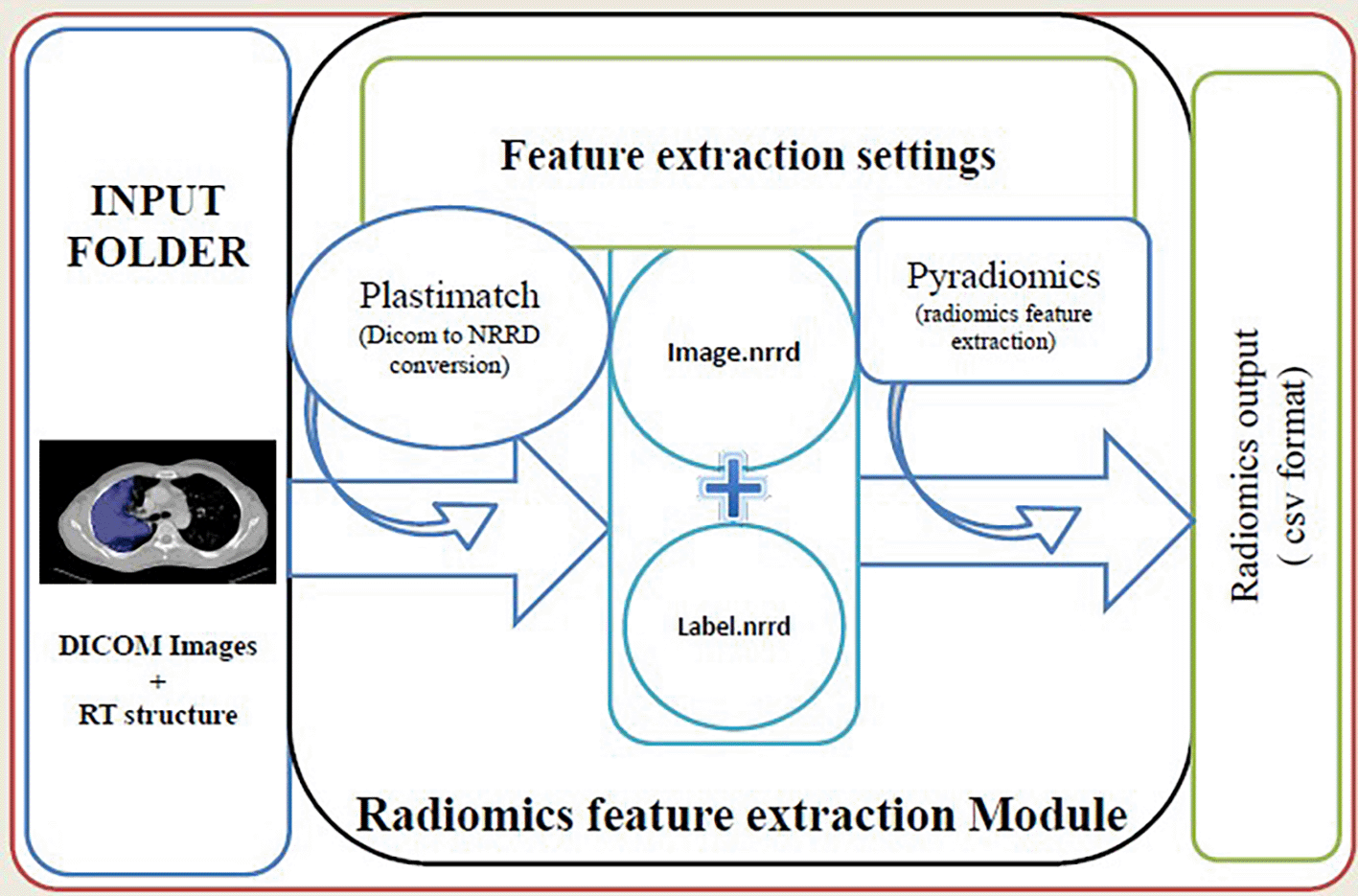
The user has to select the input folder containing the Digital Imaging and Communications in Medicine (DICOM) image and radiotherapy (RT) structure files. We can select the output folder and also customize the feature extraction process by changing values in the ‘.yaml’ file.
To start PyRadGUI, GUI_batch_radiomics.py is run on the command prompt. GUI, shown in Figure 2, has been divided into two parts, i.e., left and right containers. The left container contains three tabs that are used for customization and radiomic extraction and the right container displays the process and error if any.
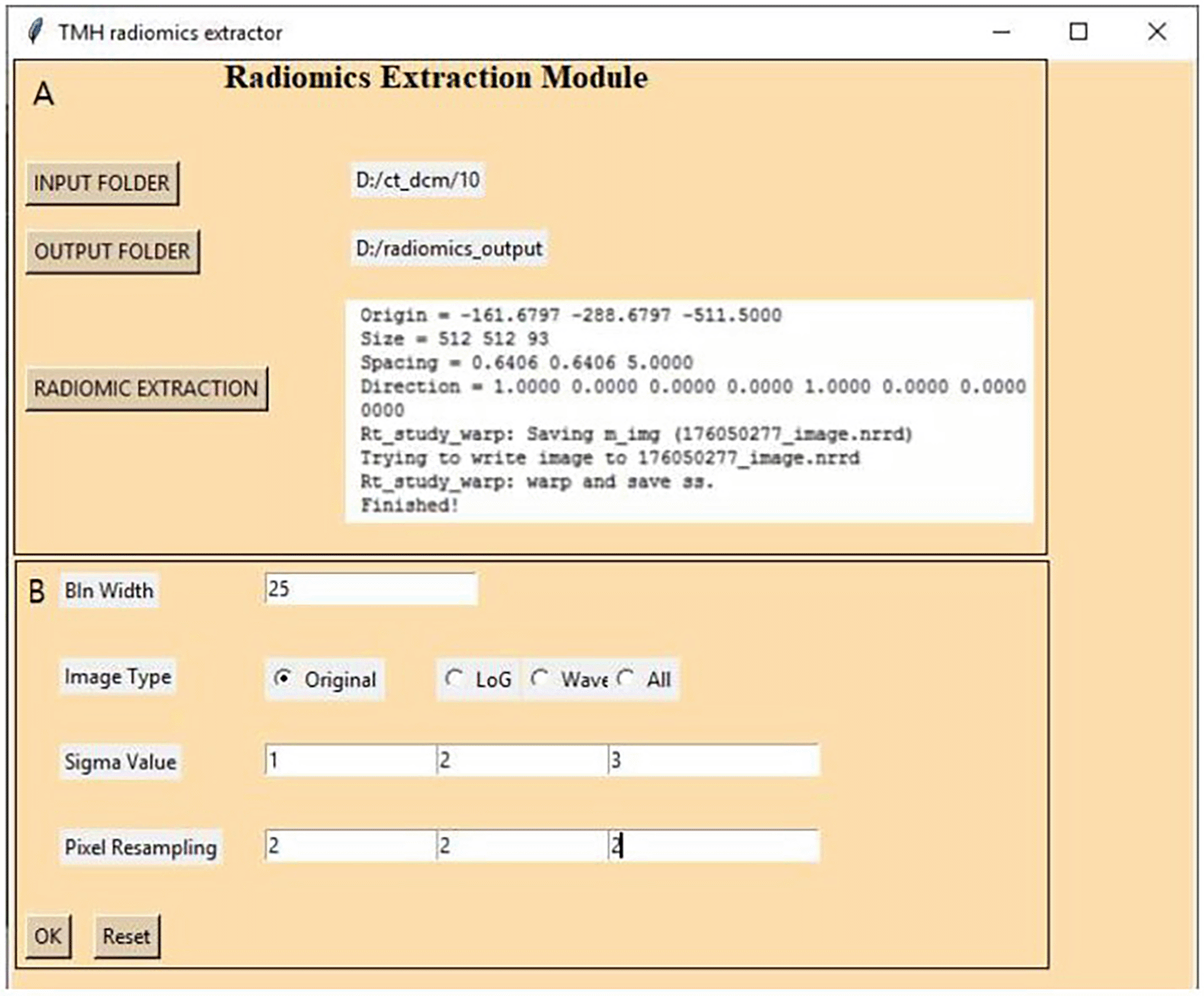
After starting the program, the user must select an input folder containing multiple patient images and a Radio therapy (RT) structure in DICOM format. Next, the user must select an output folder and then change the feature extraction settings from the settings tab.
1. Selection of image type (Original, Laplace of Gaussian, and Wavelet): To extract features on the original image, we can select the image type as the original. If feature extraction has to be done on the transformed image, then we can select either LoG (Laplace of Gaussian) or Wavelet or both.
2. Selection of feature types: We have to specify the type of features we want to extract. The default includes all 1093 features from all image types.
3. Bin width selection: In the customization window one can select bin width as per the user requirement and the default bin width is 25. We have used 25 bin widths for CT and MRI and 0.5 bin width for PET radiomic extraction.
4. Sigma value for LoG features: In the customization window one can select the sigma value for LoG features. We have selected default 1-, 2-, and 3-mm sigma values for radiomic feature extraction.
5. Selection of resampled pixel spacing: In the customization window one can select pixel spacing as desired by the user for radiomic extraction. We have used the default 2×2×2 cubic mm pixel spacing.
After the settings have been customized, radiomics extraction can be started by clicking on the radiomics extraction button. The batch extraction starts by loading the DICOM folder and calls plastimatch. Plastimatch takes the reference CT folder and converts the input DICOM to NRRD (nearly raw raster data) format. It converts the image to image.nrrd and mask to mask.nrrd. Pyradiomics takes the converted nrrd image, nrrd mask, and the settings specified by the user and then extracts the radiomics features and writes the output in CSV format in the selected output folder. In this output, CSV file columns represent radiomic features and rows represent individual patients. The first column of the file is the patient identification number. This CSV file can be used for various analyses. Status of the running of the process, success, failures, and error reports are displayed in the right container in GUI, and processing reports are stored in the output folder as log files.
We have developed and tested our software on two computer systems (machines) using various software packages. The details of system information and software packages are shown in Table 1.
In total 50 non-small cell lung carcinomas (NSCLC) patients’ PET/CT data with delineation and 20 chondrosarcoma patients’ MRI data with delineation who were imaged between 2014 to 2017 were used for validation and performance testing of this software. The patient’s demographic data is shown in Table 2. Images from 100 NSLC patients were utilized to test the batch processing.
Software validation was performed by comparing Radiomic feature value extracted using PyRadGUI Workflow (PrGW) and the two reference workflows i.e., Reference Workflow1(RW1) (3DSlicer +Pyradiomics) and Workflow2 (RW2) (Plastimatch +Pyradiomics). The same version of the Pyradiomic package and Plastimatch was used for all the workflows. For the comparison, CT imaging data from 10 patients, PET imaging data from five patients, and MRI imaging data from five patients were all employed. PyRadGUI validation algorithm workflows are shown in Figure 3.
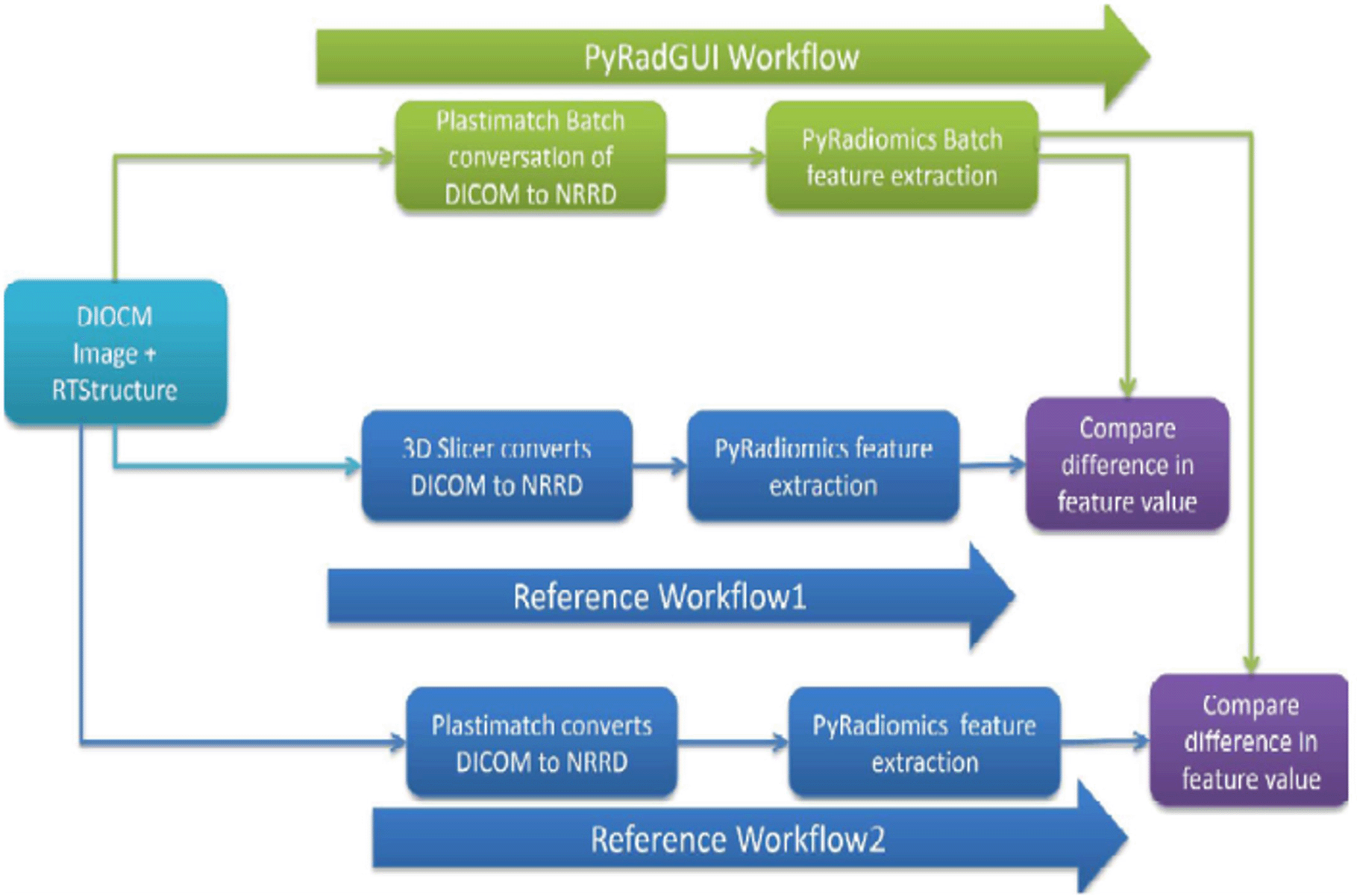
Reference Workflow1 (RW1): Individual patient’s DICOM image and ROI were loaded in 3D slicer 3.0. The first appropriateness of tumor delineation was checked by an experienced imaging physicist (15 years of experience). Image and ROI were converted in NRRD format and the image was saved as an image.nrrd and ROI as label.nrrd in the patient's image folder. Again, the NRRD image and label were loaded in 3D Slicer, and appropriateness on tumor delineation was checked by the same physicist. Subsequently, the radiomic feature was extracted and saved in CSV format using the pyradiomic package on the command prompt. The same process was repeated for all 20 patients’ data.
Reference Workflow2 (RW2): DICOM image and RTStructure were converted in NRRD format using Plastimatch 1.8.0 and stored similarly as it is done in Workflow1. Subsequently, radiomic features were extracted and stored in CSV format similarly as it is done in workflow1. The algorithm used for the manual extraction of radiomic features is shown in Figure 3. All the patient’s radiomic feature data was arranged similarly as it was arranged in the automated extraction of radiomic feature data as described in the automatic extraction section.
Our PyRadGUI workflow was compared with manual Reference Workflow1 and Reference Workflow2 as shown in Figure 3. Interclass correlation (ICC) was calculated for all 1093 radiomic features to compare PyRadGUI workflow and Reference Workflow1 using python code. As we were expecting the same result in PyRadGUI Workflow and Reference Workflow2, we used the EXACT function of Microsoft Excel 2007 software to compare both the data sets. The EXACT function compares two strings for all the characters, if all the characters are the same in both the strings it gives true as an output otherwise false.
The performance of the software was tested for batch processing on the two machines mentioned in Table 1. CT and PET DICOM data were used in the batches of 10, 20, 50, and MRI DICOM data were used in the batches of 10, and 20. The total time required for individual batch processing was recorded.
We successfully installed and ran the software on the two computers mentioned in Table 1. All the customizations for radiomic extraction work well on multiple trials. With this software, we were able to perform radiomic extraction by batch processing of up to 100 patients’ CT data on both computers.
The ICC value for the comparison of PrGW and RW1 was ICC =0.978 (range: 0.9612-1.0) across all the modalities. The ICC value for the comparison of PrGW and RW1 is shown in Table 3. All the values (for 1093 radiomic features) in the validation steps for CT, PET, and MRI features for PrGW and RW2 were TRUE, which shows that we were able to extract the same values for all the features for the same data across all the modalities by using this software as confirmed by the manual process.
On both machines the batch processing performance was found to be satisfactory. The details of run time are shown in Table 4. Various imaging modalities were used to test the PyRadGUI tool.
Several licensed and open-source software are available for radiomic extraction, which can extract radiomic features from two- or three-dimensional medical images (Gotz et al., 2019; Szczypinski et al., 2009; Zhang et al., 2015; Apte et al., 2018).
Shi et al. (2019) have developed an Ontology-guided radiomics analysis workflow (O-RAW) using the Pyradiomics package and SITK Python package, which is also able to extract radiomics features from DICOM images and RTSTRUCT in batch processing and store it in resource description framework (RDF) triple store. Another radiomic extraction software, TexRAD, is a licensed package GUI-based system. It has been used by many researchers for radiomic extraction. TexRAD is unable to do the batch processing and it can handle one patient’s data at a time (Gotz et al., 2019). This software has capabilities to display and review the image and delineate tumors manually, but another drawback of this software is its inability to use existing delineation (RTStructure) (Ganeshan et al., 2008; Davnall et al., 2012).
The RaCaT is radiomic software that implemented the IBSI defined feature but is not available as a GUI tool as well as unable to perform batch processing. The PyRadGUI software is a GUI-based tool and it can implement batch processing of DICOM images and RT structure for radiomic extraction. As this software is an extension of the pyradiomic package it inherently implements the IBSI feature definition. It can extract radiomic features from hundreds of patients’ images and RTStructure in batch processing mode and store the result in CSV format. Although we have not compared the delineation converted to NRRD using Plastimatch and 3D Slicer, radiomic feature values comparison show excellent agreement (ICC=0.998±0.012) between the two methods. As our results show, this software calculates radiomics features accurately and reliably. Radiomic extraction from PET and CT images takes a much longer time compared to MRI images, as PET and CT have whole-body images [contains more data] and MRI has regional images [contains less data]. We have used Pyradiomics, open-source software for radiomic extraction in our research infrastructure because this infrastructure can easily be replicated in other research centers. This software works as a plug-in and has no dependencies on the pyradiomic package version, it can be upgraded as and when a new pyradiomic package is available. Customization is the unique feature of this software, which provides flexibility to the user to customize the parameters in the ‘yaml’ file of the pyradiomics package. The ability of our software to customize and extract 1093 radiomic features from medical images in batch processing enables faster processing of radiomic extraction and storage of feature values in CSV format. During the customization, the user can also select a specific group of features to be extracted. The advantage of this software is its GUI and GUI-based customization of extraction, which allows performing the entire task from the GUI console by clicking the available buttons on the console. The CSV format in which this software stores data where each column represents radiomic features and rows represent individual patient’s data makes it easier to be utilized for machine learning. It can also be concatenated with clinical data if required. Log files can be used for identifying any error that occurs during the processing. We can identify the specific data that contains errors and then take corrective action by referring to the error log of each patient's data that is generated and stored in log file. In our existing project, we have also developed artificial intelligence (AI) infrastructure for AI-based research in oncology and PyRadGUI is also an integral part of it. The PyRadGUI can be implemented as standalone as well as part of AI infrastructure for radiomic based research. The portability and easy installation of this software will encourage the radiomic community to use this software and this software can be a valuable addition to radiomic based research infrastructure.
There are also a few limitations of this software like it is unable to display the image before or during the procedure. It requires a DICOM image as well as structure for radiomic processing. Future work will be to test this software on Linux operating system, add a statistical and prediction analytics module and image segmentation and display module to this tool.
We successfully implemented and validated, PyRadGUI, a GUI-based easy-to-use Radiomic extraction software. This software can easily be implemented on Windows systems. The extracted features using this software are meeting the IBSI standards. We have found this software able to perform batch processing of up to 100 patients and extract radiomic features and store it in ready-to-use CSV format for machine learning. Documentation including the description of how to install and use this software can be found on GitHub (https://github.com/Bionic-TMH/PyRadGUI).
The corresponding author has access to the data created and used in this study for verification. Data sharing on a public repository is prohibited by IRB permission. Only the project partners are permitted to access the data due to confidentiality reasons regarding patient data.
Source code available from: https://github.com/Bionic-TMH/PyRadGUI
Archived source code at time of publication: https://doi.org/10.5281/zenodo.7524822
License: Apache License 2.0
The authors wish to thank The Netherlands Enterprise Agency (RVO) and MeITy for the Indo-Dutch NWO/MeITy BIONIC and TRAIN project.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Artificial Intelligence, Brain Cancer, Statistics, Neuroscience, MRI
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Machine/Deep Learning Quantitative Imaging Radio/Genomics Neuroscience Radiomics
Is the rationale for developing the new software tool clearly explained?
Partly
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Radiomics; artificial intelligence
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 10 Mar 23 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
I read with great interest your research article about a new tool that simplify the radiomics feature extraction process. I strongly believe that we, researchers, should focus ... Continue reading Dear authors,
I read with great interest your research article about a new tool that simplify the radiomics feature extraction process. I strongly believe that we, researchers, should focus on facilitating the translation of radiomics from research to clinical practice, and the development of new user friendly tools is part of this process.
As it was correctly reported in the Introduction and in the Abstract, many software for the extraction of radiomics features exist, and only few of them are IBSI compliant, such as Pyradiomics. To complete the existing literature and state of art, I would like to cite some already developed tools that make use of pyradiomics in a user friendly enviroment.
These are:
- the SlicerRadiomics extension for 3D slicer (https://github.com/AIM-Harvard/SlicerRadiomics), that offers a gui for the pyradiomics extractor and it is embedded in 3D Slicer.
- the matRadiomics tool (https://doi.org/10.3390/jimaging8080221), a novel and complete framework that provides a friendly user interface for pyradiomics and complete the radiomics workflow adding image visualization, segmentation, feature selection and machine learning.
- FeAture Explorer (FAE) (doi:10.1371/journal.pone.0237587), that uses pyradiomics and complete the radiomics workflow with feature selection and machine learning.
My comment is not meant to diminish your work, instead I wrote it, because I believe that the diffusion of knowledge is what drives research forward.Giovanni
I read with great interest your research article about a new tool that simplify the radiomics feature extraction process. I strongly believe that we, researchers, should focus on facilitating the translation of radiomics from research to clinical practice, and the development of new user friendly tools is part of this process.
As it was correctly reported in the Introduction and in the Abstract, many software for the extraction of radiomics features exist, and only few of them are IBSI compliant, such as Pyradiomics. To complete the existing literature and state of art, I would like to cite some already developed tools that make use of pyradiomics in a user friendly enviroment.
These are:
- the SlicerRadiomics extension for 3D slicer (https://github.com/AIM-Harvard/SlicerRadiomics), that offers a gui for the pyradiomics extractor and it is embedded in 3D Slicer.
- the matRadiomics tool (https://doi.org/10.3390/jimaging8080221), a novel and complete framework that provides a friendly user interface for pyradiomics and complete the radiomics workflow adding image visualization, segmentation, feature selection and machine learning.
- FeAture Explorer (FAE) (doi:10.1371/journal.pone.0237587), that uses pyradiomics and complete the radiomics workflow with feature selection and machine learning.
My comment is not meant to diminish your work, instead I wrote it, because I believe that the diffusion of knowledge is what drives research forward.Giovanni