Keywords
Shank3, Phelan-McDermid Syndrome, autism spectrum disorders, zebrafish, Risperidone, Carbamazepine, Lithium, MPEP, anti-epileptic
Shank3, Phelan-McDermid Syndrome, autism spectrum disorders, zebrafish, Risperidone, Carbamazepine, Lithium, MPEP, anti-epileptic
This revised version of our research article is greatly improved in response to valuable reviewer feedback and provides several important clarifications. These include an explanation of "reactivity" and "activity" endpoints (in both introduction and methods sections); expanded methods describing water quality, shank3 models that include mutations in both shank3a and shank3b ohnologs; relabeling in Figures 1 & 2; and reorganization so that text and figures are integrated with tables that include all statistical analyses coming just before the references. We feel that these changes make the research more accessible and we welcome further feedback.
See the authors' detailed response to the review by Sara Moir Sarasua
See the authors' detailed response to the review by Fumihito Ono
Altered sensory processing affects the majority (69-97%) of people with autism and is one of the core diagnostic symptoms in the Diagnostic and Statistical Manual V (Leekam et al., 2007; Tomchek and Dunn, 2007; Lane et al., 2011; Green et al., 2016; Tavassoli et al., 2016; Siper et al., 2017). Such symptoms includes hypo- and hyper-reactivity to stimuli, and sensory fixation (Robertson and Baron-Cohen, 2017). Consistent with this, genotype by symptom meta-analyses identified sensory hyporeactivity/increased-pain-tolerance in over 80% of individuals with Phelan-McDermid syndrome (PMS) (Mieses et al., 2016; Tavassoli et al., 2016; De Rubeis, 2018). PMS is a syndromic form of ASD, that can be caused by a chromosome 22 terminal deletion that encompasses the SHANK3 gene or a mutation in the SHANK3 gene specifically (Phelan and McDermid, 2012; De Rubeis, 2018). In addition to sensory hyporeactivity, SHANK3 mutations are correlated with a range of symptoms, that include epilepsy, sleep disturbances, and gastrointestinal distress (Soorya et al., 2013; De Rubeis, 2018; Frank, 2021; Smith-Hicks et al., 2021). This range of symptoms makes prescribing medications challenging (Costales and Kolevzon, 2015; Harony-Nicolas et al., 2015), with many individuals experiencing a prescription carousel: when one drug fails to maintain control of a symptom and/or side-effects become intolerable. Therefore, to achieve more effective symptom management, it is critical to better understand how medications impact the range of symptoms found in individuals with PMS.
Zebrafish provide characteristics that are ideal for studying how small molecules impact sensory-motor behaviors. Zebrafish sensory-motor circuits are established and become active a few days after fertilization because precocial behavioral development is essential for the survival of freely swimming larvae (Kimmel et al., 1974; Portugues and Engert, 2009; Fero et al., 2011; Kinkhabwala et al., 2011; Warp et al., 2012; Marques et al., 2018). Predator avoidance and prey capture require visual acuity, sensitive hearing, and multimodal sensory integration to activate the appropriate swimming circuits (Fero et al., 2011; Koyama et al., 2011). Importantly, sensory-motor deficits provide a proxy for circuit pathology, that can be used to identify neuropathological critical periods (Kozol, 2018; Sakai et al., 2018; Kozol et al., 2021). Finally, due to their small size and large clutch sizes (100-200 embryos), zebrafish can be screened in large numbers and also absorb most small molecules dissolved in the water that houses them. Therefore, zebrafish provide a vertebrate model that is poised to identify how small molecules influence sensorimotor behaviors in ASD models (Sakai et al., 2018).
To investigate how drugs impact SHANK3-associated hyporeactivity, zebrafish shank3a and shank3b (shank3ab) mutants were exposed to drugs and screened for sensorimotor behavior using a the well-established visual-motor-response (VMR) assay (Burgess and Granato, 2007). During the VMR, sudden changes in illumination from light to dark evoke abrupt increases in swimming behavior as larvae search the well for a way to return to the light (Horstick et al. 2017); we capture the abrupt response by quantifying swimming in the first 30 seconds right after the transition to dark, referred to hereafter as reactivity, but the larvae sustain their search for the full 5 minutes, referred to hereafter as activity. shank3ab mutants exhibit both hyporeactivity and sustained hypoactivity in response VMR repeated lights-on to lights-off transitions (Kozol et al., 2021). To determine the effects of small molecules on this sensorimotor deficit, we exposed larval zebrafish to the commonly prescribed medications risperidone (Nyberg et al., 1993; McDougle et al., 2005; Gencer et al., 2008; Lemmon et al., 2011), lithium chloride (LiCl) (Malhi et al., 2013; Verhoeven et al., 2013; Serret et al., 2015; Egger et al., 2017; Malhi et al., 2020), and carbamazepine (CBZ) (Mattson et al., 1992; Verhoeven et al., 2013; Jia et al., 2022). We also tested 2-methyl-6-(phenylethynyl) pyridine (MPEP), which normalized anxiety and striatal synaptic transmission in a shank3 mouse model (Wang et al., 2016). Lastly, we quantified swimming before and after exposure to pentylenetetrazole (PTZ), a drug used in animal models to better understand susceptibility to seizures, at doses that normally cause hyperactivity in wild type larvae (Baraban et al., 2005; Hoffman et al., 2016; Liu and Baraban, 2019). Results of the above experiments are summarized in the column entitled ‘effect on VMR’ in Table 1.
These are based on relevant references in the rightmost column. Drug effects on VMR in are based results from this study.
| Drugs | Indication | Target(s) | Effect on VMR | Reference |
|---|---|---|---|---|
| Risperidone | Human Antipsychotic; Irritability in ASD | Various/unknown 5-HT2C; 5-HT2A; D2 a1/a2 adrenergic;H1 histamine receptor antagonists; Sodium channels | No change in WT; reduced VMR reactivity and rescued VMR sustained activity in shank3ab-/- models | (McDougle et al., 2005; Lemmon et al., 2011; Fallah et al., 2019; Panizzutti et al., 2021; Guber et al., 2022) |
| Carbamazepine CBZ | Human Anti-epileptic; Mood stabilizer | Various/unknown Sodium channels | VMR trended reduced in shank3N & WT No change in shank3C | (Mattson et al., 1992; Verhoeven et al., 2013; Jia et al., 2022) |
| LiCl | Human Mood stabilizer | Various: Dopamine; G-protein-coupled receptors; adenylate cyclase; phosphoinositide signals; MARKS, PKC, GSKb; GABA | No change in any genotype | (Malhi et al., 2013; Serret et al., 2015; Egger et al., 2017) |
| 2-Methyl-6-(phenylethynyl) pyridine MPEP | Mouse models of Fragile X, Shank3 | mGluR5 | Reduced WT VMR to shank3 levels; no change in either shank3ab-/- model | (Tu et al., 1999; Tucker et al., 2006; Vucurovic et al., 2012; Wang et al., 2016) |
| Pentylenetetrazole PTZ | Zebrafish/mouse seizure-inducing drug | GABAA receptor antagonist | Induced seizure-like activity in WT. Both shank3ab-/- models exhibit reduced response to PTZ | (Baraban et al., 2005; Dhamne et al., 2017; Liu and Baraban, 2019) |
Below we describe the varied ways these drugs impacted the VMR sensorimotor behavior, from having no effect to suppressing or enhancing the VMR in a shank3-genotype-specific manner.
Zebrafish were housed and maintained at 28°C in system-water on a 14:10 hour circadian light:dark cycle in the zebrafish core facility at the University of Miami where they were fed twice a day using a combination of dry fish food and brine shrimp. The water in which the adult fish are housed are tested for pH and conductivity by probes that are always sampling, ‘system water’. System water is tap water that goes through a water softener, a charcoal filter, and reverse osmosis membranes to make the water less hard/alkaline, remove contaminants and ions respectively. This purified water is stored on a 100 gallon storage tank and used for 10% daily water exchanges that are controlled by a solenoid. pH 7.0-8.1 and conductivity 350-800 µS are kept within range by two dosers, one with sodium bicarbonate (pH) and the other with instant ocean (conductivity). We also track room humidity and temperature on a daily basis. These values are important to track because the temperature of the water is regulated by air temperature. Adult and larval zebrafish used in this study were handled in accordance with NIH guidelines and experiments were approved by the University of Miami Institutional Care and Use Committee protocol #’s 15-128 (approval date 9/22/2015) and 18-128 (approval date 9/27/2018). To limit harm to the animals and ensure experimental reproducibility, after natural spawnings, unfertilized eggs were removed and embryos were maintained in 10 cm dishes with ~50 larvae per dish until behavioral observations. Embryos were raised with the same 14:10 light cycle as their parents. Zebrafish lines used in this study were; ABTL wildtype (WT), shank3abN-/- (Kozol et al., 2021) and shank3abC-/- (James et al., 2019). Readers should note that each model includes a mutation in both the a and the b ohnolog of the shank3 gene and therefor mutants are referred to as shank3ab; mutations in shank3abN are located near the N-terminus while those in shank3abC are located near the C-terminus of the predicted Shank3 protein product (Figure 1a).
This study is reported in line with the Animal Research: Reporting of in vivo Experiments (ARRIVE) guidelines (Kozol & Dallman, 2023).
Sample
All exact sample sizes can be found in the figure legends. Sample sizes were derived from a previous study based on the same VMR behavioral endpoint (Kozol et al., 2021).
High-throughput behavioral screens
Experimental plans were developed and refined during weekly meetings but there was no protocol registered prior to initiation of experiments. The DanioVision systemtm (Noldus, Wageningen, NTD) with the DanioVision observation chamber (DVOC-0040) was used to record videos of larval behaviors during experiments using the following settings: 25 fps, 1280 × 960 resolution using a Basler acA1300-60 gm camera fitted with a 12 mm Megapixel lens. White light for the visual motor response assay was set at 12% intensity on the high-power setting. Larvae were pipeted into an ANSI-SBS-compatible 96 well microtiter plate at a density of one larva per well, at a depth of 10 mm. Six-day-old larvae were acclimated to the observation chamber at 28 °C in the dark for at least 1 hr. Larval sex is unknown at this stage. Larvae were monitored during behavioral recordings, to ensure no signs of distress were exhibited during light cycles. DanioVision EthoVision XT software version 11.5 (Noldus) was used to set up data collection and for preliminary analyses. Visual motor response (VMR) experiments consisted of four cycles of alternating lights-on (five min.)/lights-off (five min.) for a total of 40 minutes. All behavioral experiments were conducted between 11 am and 3 pm, with 2-5 independent trials. Behavior was analyzed by binning the raw ethovision movement data into 30 second and 5 minute bins. We then defined behaviors in the first 30 seconds after dark transitions as reactivity and behaviors sustained across the full five minutes of darkness as activity. Therefore, a statistical increase or decrease in swimming during the first 30 seconds was defined as hyperreactive or hyporeactive respectively; a statistical increase or decrease in swimming during the full five minutes was defined as hyperactive or hypoactive respectively. Larvae were randomly assigned across each 96-well plate, blinded to experimenters, then were genotyped following behavioral experiments using restriction digest assays previously described (James et al., 2019; Kozol et al., 2021), allowing larvae to be binned by genotype for subsequent analyses. Following experiments, larvae were humanely euthanized using MS222 (200 mg/L dissolved in system water).
Drug screening
Zebrafish were exposed to drugs dissolved in 0.1% DMSO system water (water from the system that houses the adult fish) 24 hours prior to running VMR assays. A range of risperidone, MPEP, CBZ and LiCl concentrations were derived from previously published papers (Tucker et al., 2006; Bruni et al., 2016; Hoffman et al., 2016), then dose-response curves were generated to determine an effective dose in relation to the VMR response of WT zebrafish. Concentrations used for comparing WT and shank3 larvae were 10 μM Risperidone (Bruni et al., 2016; Hoffman et al., 2016), 5 mM LiCl and 200 μM CBZ, and 5 μM MPEP (Tucker et al., 2006). Genotype controls were exposed to DMSO (0.1%) in system water.
For PTZ trials, larvae were initially acclimated in 1 mL of system water at 28 °C in the Daniovision behavioral box for 30 minutes. Larvae were then recorded for 10 minutes to establish baseline behavior. Following a baseline recording, larvae were either exposed to 3 mM PTZ in 0.1% DMSO system water or 0.1% DMSO system water for ten minutes, before capturing ten minutes of behavior following drug exposure. Baseline and PTZ/DMSO data was then binned as total distance moved for 10 minutes pre and post PTZ exposure. Both heterozygote and homozygote larvae were tested for seizure susceptibility, however to remain consistent with the other genotypes analyzed in the study, we chose to focus on the homozygote data.
Data were analyzed using PRISM 9 (graphpad, inc.); these same analyses could be conducted using R. Videos were manually screened before running data analyses, to determine that tracking software accurately captured individuals’ movements; if discrepancies between tracks and videos were noted, videos were retracked. No individuals or data points were excluded from behavioral analyses. Significance was assessed using the non-parametric Wilcoxon rank score test (Mann-Whitney rank scores). When there were more than two groups, a Kruskal-Wallis rank score test was first calculated and, if p<0.05, was followed by a Dunn’s multiple comparisons test to compare all treatments and genotypes. See Tables 2-41.
We previously showed that both shank3abN and shank3abC mutants exhibit sensory hyporeactivity (activity during first 30 seconds in dark) and hypoactivity (activity over full 5 minutes in dark) in a light to dark transition paradigm, the VMR assay (Kozol et al., 2021). Here we repeat this assay, but this time in the presence of the drug carrier 0.1% DMSO. In comparison to WT (Figure 1a & b, Tables 2 & 3), both shank3abN-/- and shank3abC-/- models exhibited hyporeactivity and hypoactivity (Figure 1c-e, Tables 4-7). These results provide a reliable sensorimotor phenotype that can be quantified following exposure to selected drugs (Kozol & Dallman, 2023).
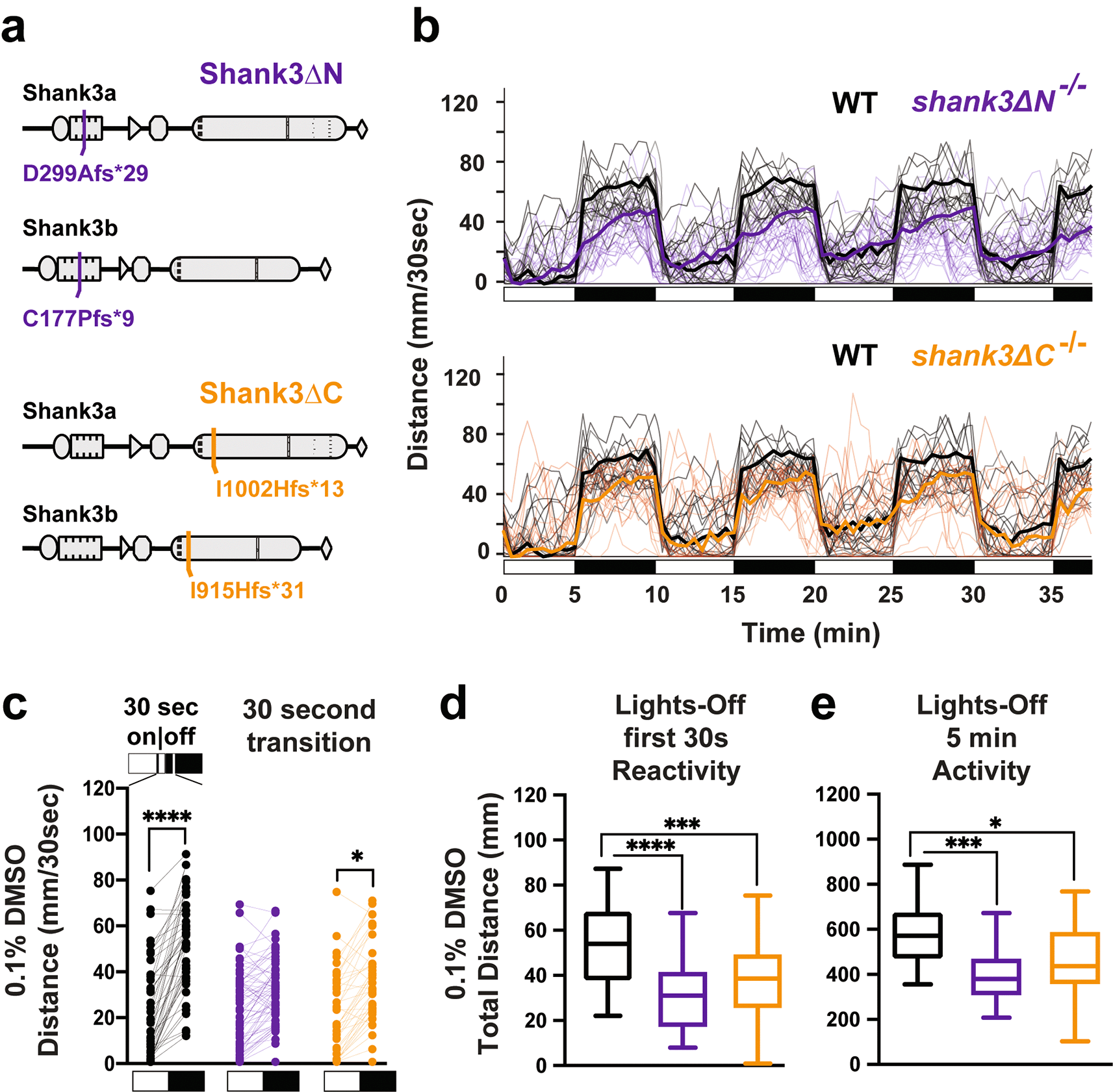
a) shank3ab N-terminal and C-terminal mutants were designed to target regions with known deleterious mutations in individuals with PMS. b) Trace line graphs showing four cycles of 5 minutes lights-on to lights-off. Checkered boxes on the x-axis represent lights on and off. c) Lights on to off paired comparison, highlighting no significant change in activity of shank3ab N terminal mutants during the first 30 sec lights-off. d) Box plots showing first 30 sec lights-off activity. e) Box plots showing activity across the full 5 minutes lights-off. Box plots represent 25th and 75th percentile, and median, with min to max whiskers. Sample sizes: WT = 50, shank3 N = 65, shank3 C = 44. p values; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Dose-response curves for small molecules were performed to investigate how these drugs impact the VMR in WT larvae. Risperidone did not affect the VMR at 1 μM, while at 10 and 20 μM doses, the VMR was decreased (Figure 2a, Tables 8-11). LiCl did not impact the VMR in WT larvae, despite exceeding previously published concentrations (Figure 2b, Tables 12-13). In contrast, CBZ had varying effects on both reactivity and activity: 80 μM and 120 μM CBZ concentrations showed no effect a, while 200 μM caused larvae to be hypo-reactive (Figure 2c, Tables 14-17). Similarly, 1 μM of MPEP did not affect the VMR, while 5 and 10 μM the VMR was decreased (Figure 2d, Tables 18-21). These results provide the lowest effective concentrations for each drug, risperidone (10 μM), CBZ (200 μM) and MPEP (5 μM), that caused a significant decrease in WT activity and reactivity; for LiCl we proceeded with the high dose of 5 mM. We next used these small molecule concentrations to compare how each would impact sensorimotor behavior in shank3ab-/- mutants.
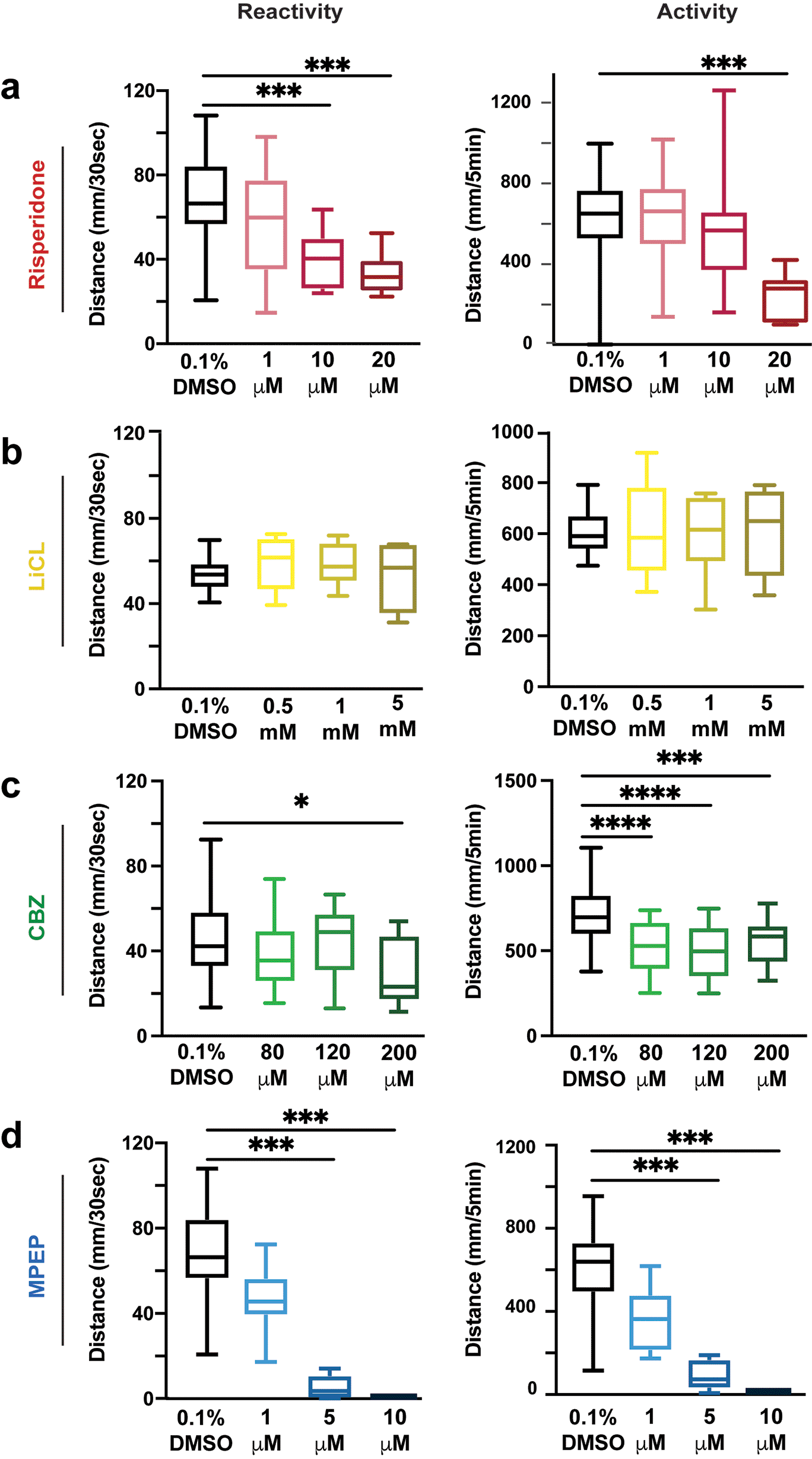
a) Risperidone exposure of WT larvae in 1, 10 and 20 μM doses. b) LiCl salt exposure of 0.5, 1 and 5 mM doses. c) CBZ exposure of WT larvae in 80, 120 and 200 μM doses. d) MPEP exposure of WT larvae in 1, 5 and 10 μM doses. Box plots represent 25th and 75th percentile, and median, with min to max whiskers. Sample sizes: WT = 23, WT + risperidone = 24, shank3 N = 33, shank3 N + risperidone = 31, shank3 C = 19, shank3 C + risperidone = 23. p values; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Risperidone is commonly prescribed in ASD for aggressive, self-injurious and hyperactive behavior (Lemmon et al., 2011). In shank3ab-/- mutants, 10 μM risperidone exacerbated hyporeactivity, but normalized hypoactivity, with shank3ab mutants achieving wild-type levels of swimming over the full duration of lights-off conditions (Figure 3, Tables 22-25). These results show that risperidone both reduced shank3 stimulus reactivity, and normalized overall stimulus-driven behaviors in shank3ab mutants.
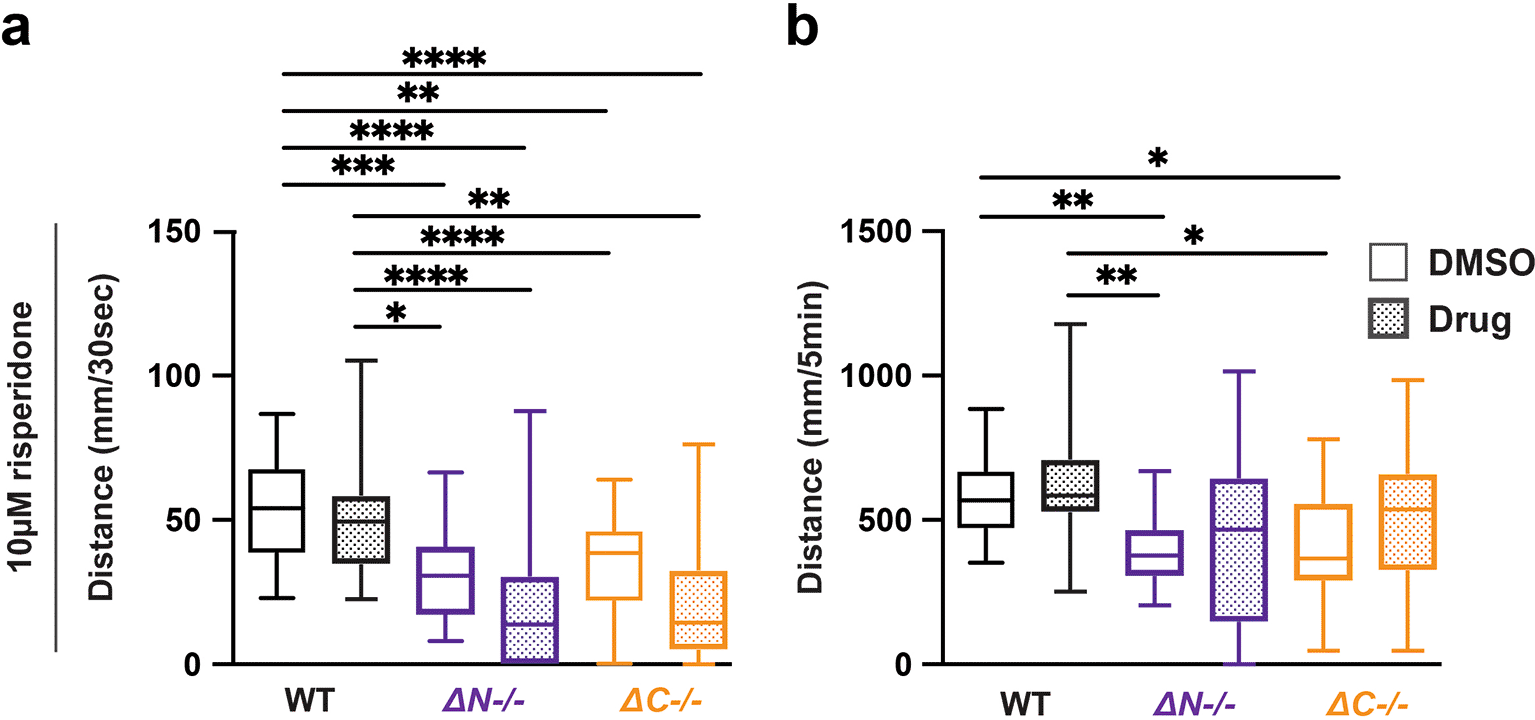
a) Activity during the first 30 seconds of lights-off of larvae exposed to 10 μM risperidone. b) Activity during the full 5 minutes lights-off of larvae exposed to 10 μM risperidone. Box plots represents 25th and 75th percentile, and median, with min to max whiskers. Sample sizes: WT = 31, WT + risperidone = 29, shank3 N = 24, shank3 N + risperidone = 23, shank3 C = 25, shank3 C + risperidone = 23. p values; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Lithium chloride (LiCl) has been prescribed for several neuropsychological disorders, including bipolar disorder, depression, and ASD (Malhi et al., 2020). LiCl has been prescribed to individuals with PMS that exhibit bipolar depression, psychosis, and catatonic behavior (Verhoeven et al., 2013; Egger et al., 2017). Exposure to 5 mM LiCl caused no change in shank3ab-/- VMR (Figure 4, Tables 26-29). Therefore, LiCl does not impact visual processing in either WT zebrafish or shank3 mutant larvae.
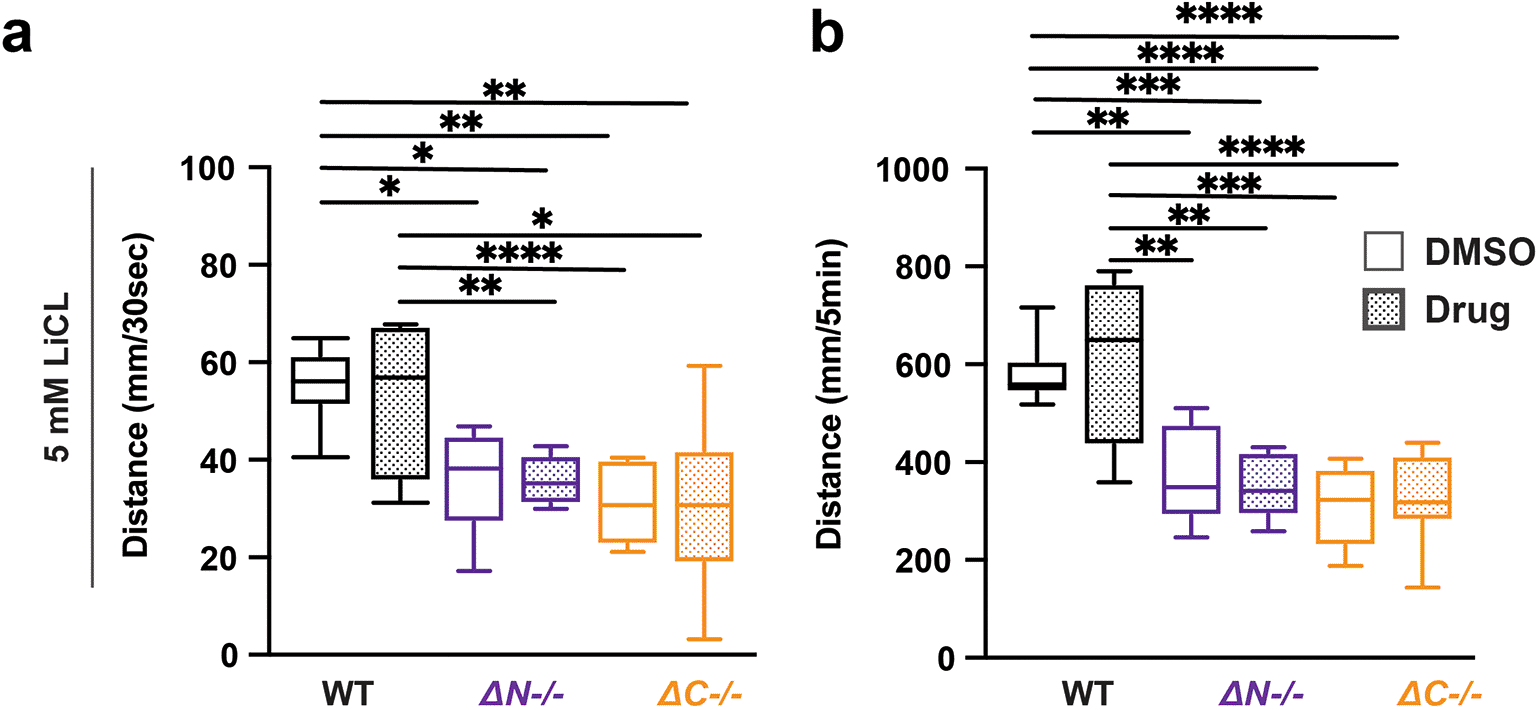
a) Activity during the first 30 seconds of lights-off of larvae exposed to 5 mM LiCl. b) Activity during the full 5 minutes lights-off of larvae exposed to 5 mM LiCl. Box plots represent 25th and 75th percentile, and median, with min to max whiskers. Sample sizes: WT = 20, WT + risperidone = 16, shank3 N = 18, shank3 N + risperidone = 18, shank3 C = 16, shank3 C + risperidone = 16. p values; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Carbamazepine (CBZ) is commonly prescribed to control seizures in individuals with epilepsy (Mattson et al., 1992). For individuals with PMS, CBZ has been prescribed following symptom resistance to common mood stabilizers, such as lithium and valproic acid (Verhoeven et al., 2013). WT and shank3abN-/- mutants VMR reactivity trended reduced with CBZ exposure but did not reach p < 0.05 (Figure 5, Tables 30-33). By contrast, shank3ab C-terminal VMR reactivity was unaffected by CBZ exposure. These results suggest that CBZ could have differential impacts on sensorimotor circuits depending on the location of the mutation in the shank3 gene.
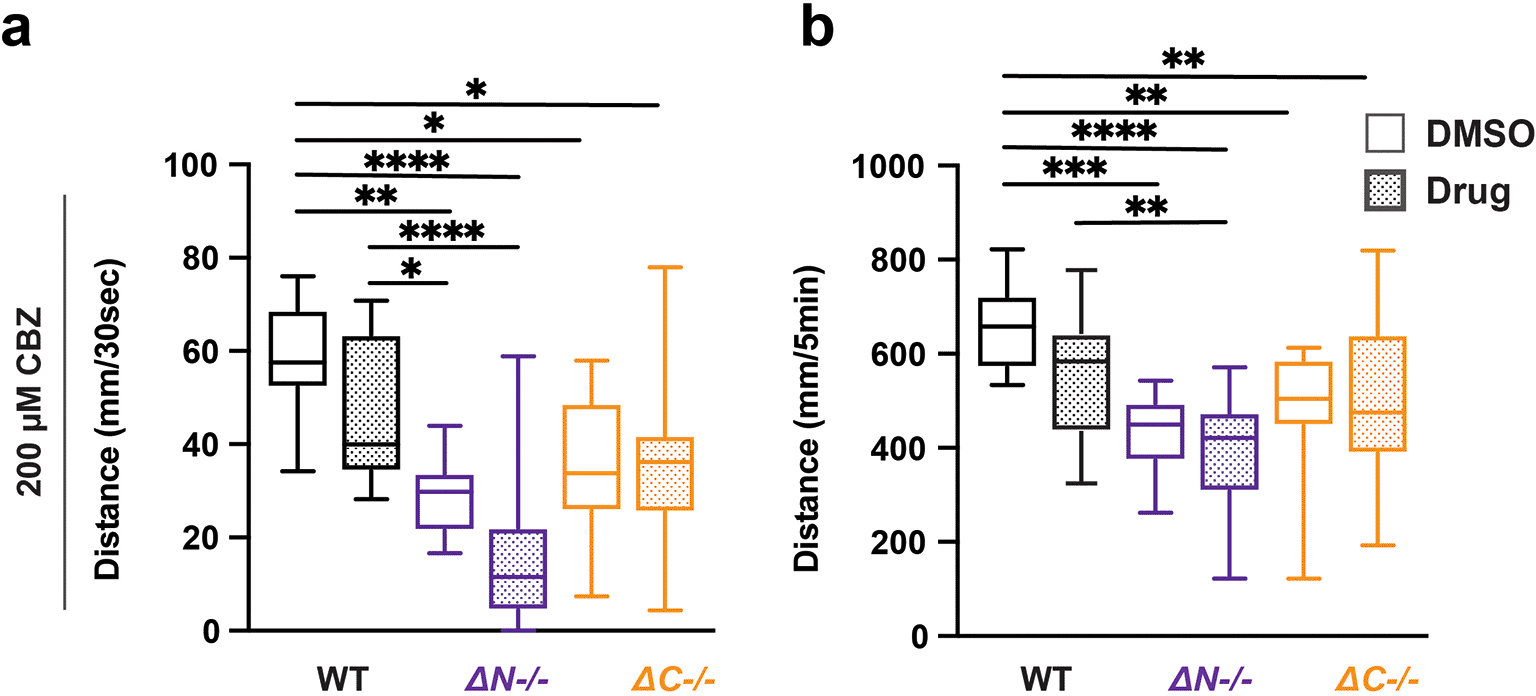
a) Activity during the first 30 seconds of lights-off of larvae exposed to 200 μM CBZ. b) Activity during the full 5 minutes lights-off of larvae exposed to 200 μM CBZ. Box plots represent 25th and 75th percentile, and median, with min to max whiskers. Sample Sizes, WT = 35, WT + CBZ = 37, shank3ab N = 24, shank3ab N + CBZ = 19, shank3ab C = 27, and shank3ab C + CBZ = 34. p values; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
While the molecules described above have been prescribed for ASD and epilepsy, we were also interested in investigating compounds used to rescue behavioral deficits in Shank3 mouse models (Wang et al., 2016). We found that MPEP did not affect the VMR in shank3ab mutants however, MPEP was sufficient to cause hyporeactivity and hypoactivity in WT larvae (Figure 6, Tables 34-37). Therefore effects of MPEP on sensory-induced behaviors were genotype-dependent.
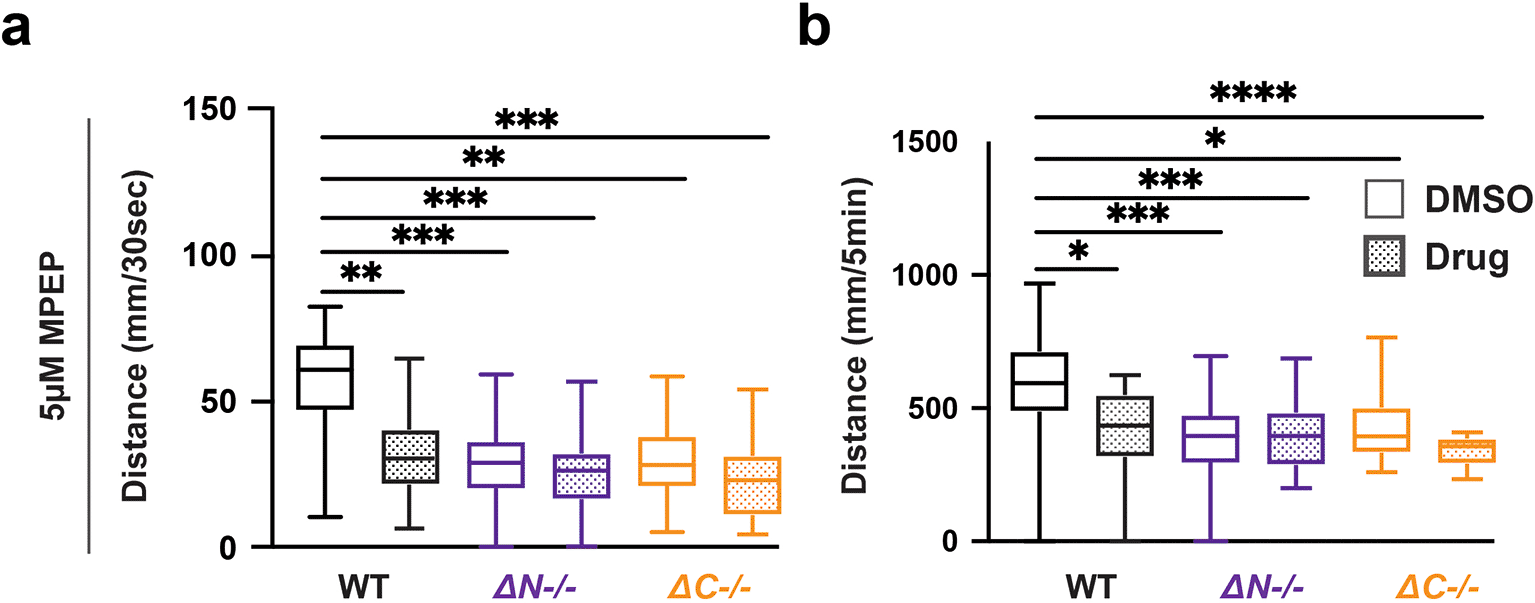
a) Activity during the first 30 seconds of lights-off of larvae exposed to 5 μM MPEP. b) Activity during the full 5 minutes lights-off of larvae exposed to 5 μM MPEP. Box plots represent 25th and 75th percentile, and median, with min to max whiskers. Sample Sizes, WT = 23, WT + MPEP = 24, shank3ab N = 33, shank3ab N + MPEP = 31, shank3ab C = 19, and shank3ab C + MPEP= 24. p values; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
A standard approach used in animal models to test for susceptibility to seizures related to reduced GABAergic inhibition is to test responses to the GABAA receptor antagonist pentylenetetrazole (PTZ) (Baraban et al., 2005; Hoffman et al., 2016; Liu and Baraban, 2019). In response to 3 mM PTZ, both N and C shank3ab-/- larvae fail to exhibit WT level of hyperactivity suggesting altered GABAergic signaling in the shank3ab mutant models (Figure 7, Tables 38-41).
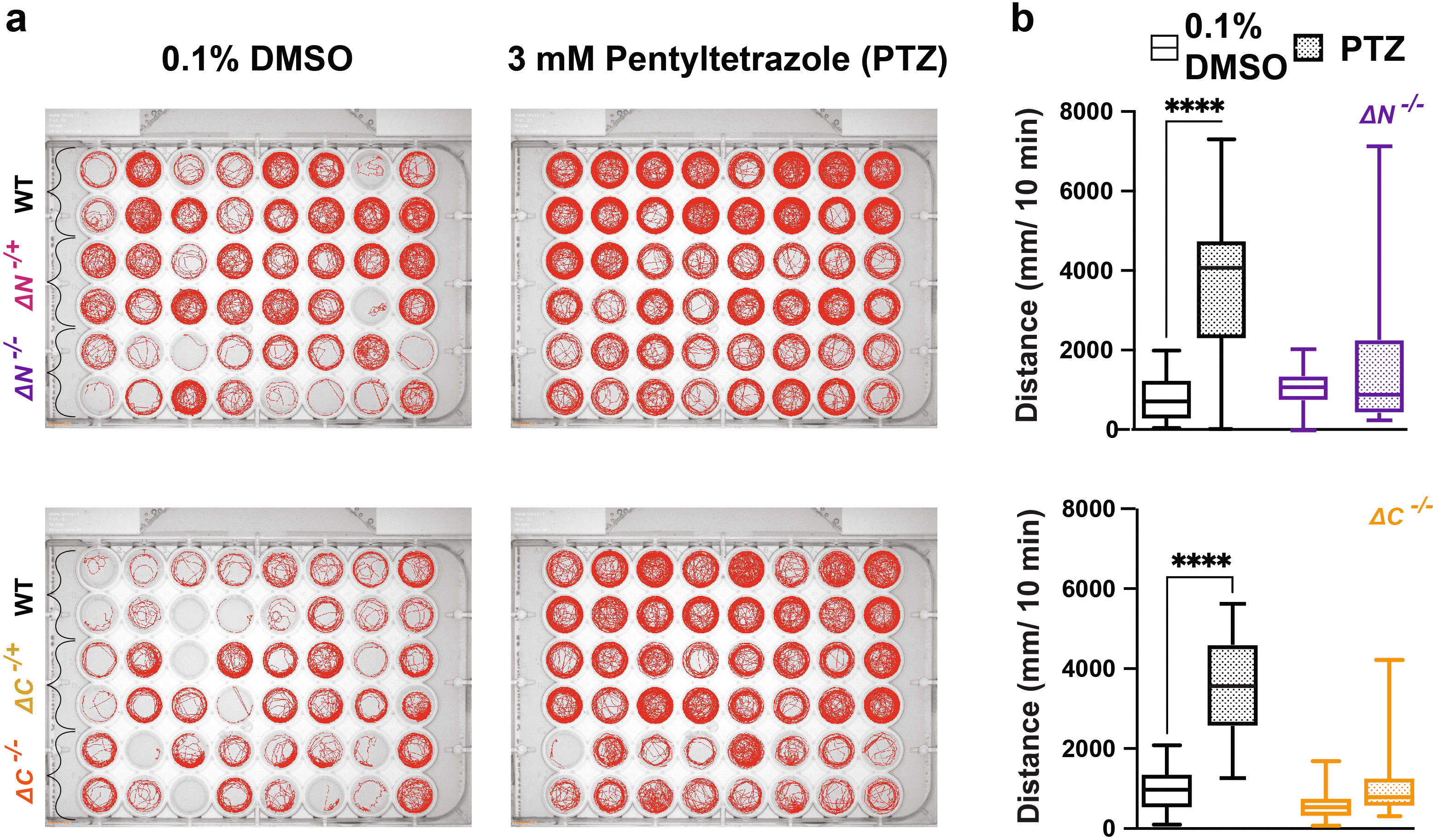
a) Behavioral traces of larvae exposed to 3 mM PTZ. b) Activity of WT, shank3ab N and shank3ab C larvae for 10 minutes following exposure to 3 mM of PTZ. Box plots: box represents 25th and 75th percentile, and median, with min to max whiskers. Sample sizes for shank3 N trials, WT = 30, shank3 N = 30; for shank3 C trials, WT = 31, shank3 C = 28. p values; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Here we show both drug- and genotype- specific effects on sensory-evoked VMR behavior in shank3ab zebrafish models of Phelan-McDermid Syndrome. The array of symptoms experienced by people with Phelan-McDermid Syndrome is likely a consequence of the diverse developmental and physiological roles played by SHANK3 (Sheng and Kim, 2000; Grabrucker, 2014; Harony-Nicolas et al., 2015; Kozol et al., 2015, 2021; Harris et al., 2016; Engineer et al., 2018; James et al., 2019; Breen et al., 2020; Lutz et al., 2020). Here we tested how drugs targeting aggressive behavior, catatonia, and/or epilepsy affect sensorimotor VMR behaviors in zebrafish shank3 models of PMS. We found that drugs were neutral, enhanced or suppressed sensory-induced behavior in a genotype- and drug-dependent manner.
Zebrafish, in particular, provide a cost-effective and high-throughput way to test how medications impact behaviors (Rihel et al., 2010; Kokel and Peterson, 2011; Rihel and Schier, 2013; Jordi et al., 2015; Bruni et al., 2016; Hoffman et al., 2016). We previously validated shank3ab N and C zebrafish models and showed a shank3ab mutant dose-dependent reduction in the VMR (James et al., 2019; Kozol et al., 2021). Because the VMR phenotype is strongest in shank3ab homozygous larvae, we focused on this genotype for our small drug screen.
Widely-prescribed, mood-stabilizing medications risperidone and LiCl had distinct effects on the VMR. Risperidone exacerbated shank3 VMR hyporeactivity and rescued overall activity to WT levels; by contrast, LiCl had no effect on the VMR in any of the three genotypes tested. In addition to the beneficial effects of risperidone however, this medication is associated with weight-gain in humans and reduced gastrointestinal motility in zebrafish (de Alvarenga et al., 2017; Guber et al., 2022). Consistent with this, risperidone D2 and 5-HT2 receptor targets (Nyberg et al., 1993) are expressed and regulate function in both brain and gut (Taniyama et al., 2000; Eliassi et al., 2008; Feng et al., 2020). Therefore, risperidone creates known symptom trade-offs in addition to improving mood in people and visual processing in zebrafish.
Treatment-resistant epilepsy in Phelan-McDermid Syndrome is one of the most difficult symptoms to manage and also one for which there are many drug options (Chakraborty et al., 2022). CBZ, a sodium channel blocker, has been used in patients with PMS who were resistant to mood stabilizers (Mattson et al., 1992; Verhoeven et al., 2013, 2020). CBZ reduced reactivity to dark transitions in WT and shank3abN-/- larvae (though VMRs in neither genotype reached p<0.05) but had no effect on median VMR values in shank3abC-/- larvae, indicating possible shank3 allele-specific differences in the way CBZ impacts the VMR. Consistent with shank3 allele-specific differences, whole brain activity mapping in these same models showed a greater activity in mid and hindbrain circuits in response to dark transition in shank3abN than shank3abC alleles (Kozol et al., 2021). Another drug that addresses seizure susceptibility is PTZ, a GABAA receptor antagonist that is used to test seizure susceptibility in zebrafish and murine models. Our findings that shank3ab models are resistant to doses that make WT larvae hyperactive suggest that these models might have fewer GABAA receptors targets for PTZ to act upon. As with the mood stabilizers, the effects of CBZ and PTZ were both drug- and genotype-dependent.
Finally, our findings that MPEP made WT behave like shank3ab-/- larvae in the VMR assay suggest that blocking mGluR5 may affect sensory processing. MPEP blocks mGluR5 and improves excessive grooming and striatal synaptic plasticity in a mouse shank3 model (Wang et al., 2016). GluR5 continues to show promise as a regulator of excitatory/inhibitory balance in the striatum where a negative correlation between mGluR5 and GABA was measured in autistic people using fMRI; mouse Cntnap2 mutants showed a similar negative mGluR5 and GABA correlation that was not found in either Shank3 or 16p11.2 deletion models (Carey et al., 2022).
Our findings highlight the genotype-, drug-, and phenotype-specific challenges of designing treatment strategies for Phelan-McDermid Syndrome. These include trade-offs that can occur when a drug like risperidone improves sensory-processing and mood at the expense of gut function and differential effects of drugs on different symptoms.
See Figure 1c.
| Table analyzed | DMSO 30sec paired | ||||
|---|---|---|---|---|---|
| Two-way ANOVA | Ordinary | ||||
| Alpha | 0.05 | ||||
| Source of Variation | % of total variation | P value | P value summary | Significant? | |
| Interaction | 4.261 | 0.0017 | ** | Yes | |
| Row Factor Light | 5.139 | 0.0005 | *** | Yes | |
| Column Factor genotype | 17.21 | <0.0001 | **** | Yes | |
| ANOVA table | SS (Type III) | DF | MS | F (DFn, DFd) | P value |
| Interaction | 4421 | 2 | 2211 | F (2, 214) = 6.588 | P=0.0017 |
| Row Factor genotype | 5333 | 2 | 2666 | F (2, 214) = 7.946 | P=0.0005 |
| Column Factor light | 17854 | 1 | 17854 | F (1, 214) = 53.20 | P<0.0001 |
| Residual | 71814 | 214 | 335.6 | ||
| Difference between column means | |||||
| Predicted (LS) mean of Lights-on | 21.82 | ||||
| Predicted (LS) mean of Lights-off | 40.23 | ||||
| Difference between predicted means | -18.41 | ||||
| SE of difference | 2.523 | ||||
| 95% CI of difference | -23.38 to -13.43 | ||||
| Data summary | |||||
| Number of columns (Light) | 2 | ||||
| Number of rows (genotype) | 3 | ||||
| Number of values | 220 |
See Figure 1c.
| Paired Comparison Lights-on to Lights-off | ||||||||
|---|---|---|---|---|---|---|---|---|
| Number of families | 1 | |||||||
| Number of comparisons per family | 3 | |||||||
| Alpha | 0.05 | |||||||
| Bonferroni's multiple comparisons test | Predicted (LS) mean diff. | 95.00% CI of diff. | Below threshold? | Summary | Adjusted P Value | |||
| Lights-on - Lights-off | ||||||||
| WT | -30.24 | -39.46 to -21.02 | Yes | **** | <0.0001 | |||
| shank3abN-/- | -9.882 | -20.93 to 1.169 | No | ns | 0.0962 | |||
| shank3abC-/- | -15.09 | -26.34 to -3.843 | Yes | ** | 0.0042 | |||
| Test details | Predicted (LS) mean 1 | Predicted (LS) mean 2 | Predicted (LS) mean diff. | SE of diff. | N1 | N2 | t | DF |
| Lights-on - Lights-off | ||||||||
| WT | 22.31 | 52.55 | -30.24 | 3.82 | 46 | 46 | 7.917 | 214 |
| shank3abN-/- | 21.11 | 30.99 | -9.882 | 4.58 | 32 | 32 | 2.158 | 214 |
| shank3abC-/- | 22.04 | 37.13 | -15.09 | 4.662 | 38 | 26 | 3.237 | 214 |
See Figure 1d.
| Table analyzed | First 30 sec Off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.001 |
| Exact or approximate P value? | Approximate |
| P value summary | *** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 3 |
| Kruskal-Wallis statistic | 54.04 |
| Data summary | |
| Number of treatments (columns) | 3 |
| Number of values (total) | 159 |
See Figure 1d.
| Number of families | 1 | |||||
| Number of comparisons per family | 3 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT vs. shank3abN-/- | 61.85 | Yes | *** | <0.001 | A-B | |
| WT vs. shank3abC-/- | 49.2 | Yes | *** | <0.001 | A-C | |
| shank3abN vs. shank3abC | -12.66 | No | ns | 0.48 | B-C | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. shank3abN-/- | 118.9 | 57.05 | 61.85 | 50 | 65 | 7.142 |
| WT vs. shank3abC-/- | 118.9 | 69.7 | 49.2 | 50 | 44 | 5.169 |
| shank3abN vs. shank3abC | 57.05 | 69.7 | -12.66 | 65 | 44 | 1.408 |
See Figure 1e.
| Table analyzed | 5 min Off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.001 |
| Exact or approximate P value? | Approximate |
| P value summary | *** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 3 |
| Kruskal-Wallis statistic | 40.54 |
| Data summary | |
| Number of treatments (columns) | 3 |
| Number of values (total) | 159 |
See Figure 1e.
| Number of families | 1 | |||||
| Number of comparisons per family | 3 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT vs. shank3abN-/- | 53.78 | Yes | *** | <0.001 | A-B | |
| WT vs. shank3abC-/- | 41.9 | Yes | *** | <0.001 | A-C | |
| shank3abN vs. shank3abC | -11.88 | No | ns | 0.56 | B-C | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. shank3abN-/- | 113.6 | 59.8 | 53.78 | 50 | 65 | 6.209 |
| WT vs. shank3abC-/- | 113.6 | 71.68 | 41.9 | 50 | 44 | 4.402 |
| shank3abN vs. shank3abC | 59.8 | 71.68 | -11.88 | 65 | 44 | 1.322 |
See Figure 2a.
| Table analyzed | Wildtype risperidone dose response 30 sec Lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups [Risperidone] | 4 |
| Kruskal-Wallis statistic | 24.09 |
| Data summary | |
| Number of treatments (columns) | 4 |
| Number of values (total) | 114 |
See Figure 2a.
| Number of families | 1 | |||||
| Number of comparisons per family | 6 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT vs. 1 uM Risp | 10.89 | No | ns | >0.9999 | A-B | |
| WT vs. 10 uM RIsp | 26.1 | Yes | ** | 0.0015 | A-C | |
| WT vs. 20 uM Risp | 48.19 | Yes | *** | 0.0003 | A-D | |
| 1 uM Risp vs. 10 uM RIsp | 15.22 | No | ns | 0.753 | B-C | |
| 1 uM Risp vs. 20 uM Risp | 37.3 | Yes | * | 0.0406 | B-D | |
| 10 uM RIsp vs. 20 uM Risp | 22.08 | No | ns | 0.4381 | C-D | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. 1 uM Risp | 71.08 | 60.19 | 10.89 | 53 | 16 | 1.155 |
| WT vs. 10 uM RIsp | 71.08 | 44.97 | 26.1 | 53 | 36 | 3.657 |
| WT vs. 20 uM Risp | 71.08 | 22.89 | 48.19 | 53 | 9 | 4.044 |
| 1 uM Risp vs. 10 uM RIsp | 60.19 | 44.97 | 15.22 | 16 | 36 | 1.532 |
| 1 uM Risp vs. 20 uM Risp | 60.19 | 22.89 | 37.3 | 16 | 9 | 2.708 |
| 10 uM RIsp vs. 20 uM Risp | 44.97 | 22.89 | 22.08 | 36 | 9 | 1.793 |
See Figure 2a.
| Table analyzed | Wildtype risperidone dose response 5 min Lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups [risperidone] | 4 |
| Kruskal-Wallis statistic | 24.89 |
| Data summary | |
| Number of treatments (columns) | 4 |
| Number of values (total) | 115 |
See Figure 2a.
| Number of families | 1 | |||||
| Number of comparisons per family | 6 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT vs. 1 uM | 3.684 | No | ns | >0.9999 | A-B | |
| WT vs. 10 uM | 16.41 | No | ns | 0.1362 | A-C | |
| WT vs. 20 uM | 55.03 | Yes | **** | <0.0001 | A-D | |
| 1 uM vs. 10 uM | 12.72 | No | ns | >0.9999 | B-C | |
| 1 uM vs. 20 uM | 51.35 | Yes | *** | 0.0008 | B-D | |
| 10 uM vs. 20 uM | 38.63 | Yes | ** | 0.0071 | C-D | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. 1 uM | 68.43 | 64.75 | 3.684 | 53 | 16 | 0.3873 |
| WT vs. 10 uM | 68.43 | 52.03 | 16.41 | 53 | 36 | 2.278 |
| WT vs. 20 uM | 68.43 | 13.4 | 55.03 | 53 | 10 | 4.788 |
| 1 uM vs. 10 uM | 64.75 | 52.03 | 12.72 | 16 | 36 | 1.27 |
| 1 uM vs. 20 uM | 64.75 | 13.4 | 51.35 | 16 | 10 | 3.821 |
| 10 uM vs. 20 uM | 52.03 | 13.4 | 38.63 | 36 | 10 | 3.241 |
See Figure 2b.
See Figure 2b.
See Figure 2c.
| Table analyzed | Wildtype CBZ dose response 30 sec Lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | 0.0422 |
| Exact or approximate P value? | Approximate |
| P value summary | * |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups [CBZ] | 4 |
| Kruskal-Wallis statistic | 8.191 |
| Data summary | |
| Number of treatments (columns) | 4 |
| Number of values (total) | 97 |
See Figure 2c.
| Number of families | 1 | |||||
| Number of comparisons per family | 3 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | A-? | |
| WT vs. WT 80 uM CBZ | 8.93 | No | ns | 0.8191 | B | WT CBZ |
| WT vs. WT 120 uM CBZ | 1.646 | No | ns | >0.9999 | C | WT CBZ 2 |
| WT vs. WT CBZ 200 uM CBZ | 22 | Yes | * | 0.0172 | D | WT CBZ 3 |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. WT 80 uM CBZ CBZ | 54.62 | 45.69 | 8.93 | 47 | 16 | 1.096 |
| WT vs. WT 120 uM CBZ CBZ | 54.62 | 52.97 | 1.646 | 47 | 17 | 0.2067 |
| WT vs. WT 200 uM CBZ CBZ | 54.62 | 32.62 | 22 | 47 | 17 | 2.762 |
See Figure 2c.
| Table analyzed | Wildtype CBZ 5 min Lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 4 |
| Kruskal-Wallis statistic | 28.11 |
| Data summary | |
| Number of treatments (columns) | 4 |
| Number of values (total) | 97 |
See Figure 2c.
| Number of families | 1 | |||||
| Number of comparisons per family | 3 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | A-? | |
| WT vs. WT 80 uM CBZ | 30.76 | Yes | *** | 0.0005 | B | WT CBZ |
| WT vs. WT 120 uM CBZ CBZ | 32.83 | Yes | *** | 0.0001 | C | WT CBZ |
| WT vs. 20 uM CBZ WT cbz | 26.72 | Yes | ** | 0.0024 | D | WT cbz |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. WT 80 uM CBZ | 64.51 | 33.75 | 30.76 | 47 | 16 | 3.776 |
| WT vs. WT 120 uM CBZ | 64.51 | 31.68 | 32.83 | 47 | 17 | 4.122 |
| WT vs. WT 200 uM CBZ | 64.51 | 37.79 | 26.72 | 47 | 17 | 3.354 |
See Figure 2d.
| Table analyzed | Wildtype MPEP 30 sec Lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 4 |
| Kruskal-Wallis statistic | 50.85 |
| Data summary | |
| Number of treatments (columns) | 4 |
| Number of values (total) | 68 |
See Figure 2d.
| Number of families | 1 | |||||
| Number of comparisons per family | 3 | |||||
| Alpha | 0.05 | |||||
| Dunn’s multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | A-? | |
| WT vs. WT 1 uM MPEP | 12.47 | No | ns | 0.1319 | B | WT 1 uM MPEP |
| WT vs. WT 5 uM MPEP | 33.87 | Yes | **** | <0.0001 | C | WT 5 uM MPEP |
| WT vs. WT 10 uM MPEP | 41.34 | Yes | **** | <0.0001 | D | WT 10 uM MPEP |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. WT 1 uM MPEP | 51.04 | 38.56 | 12.47 | 28 | 16 | 2.014 |
| WT vs. WT 5 uM MPEP | 51.04 | 17.17 | 33.87 | 28 | 9 | 4.474 |
| WT vs. WT 10 uM MPEP | 51.04 | 9.7 | 41.34 | 28 | 15 | 6.538 |
See Figure 2d.
| Table analyzed | Wildtype MPEP dose response 5 min Lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 4 |
| Kruskal-Wallis statistic | 50.52 |
| Data summary | |
| Number of treatments (columns) | 4 |
| Number of values (total) | 66 |
See Figure 2d.
| Number of families | 1 | |||||
| Number of comparisons per family | 3 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | A-? | |
| WT vs. WT 1 μM MPEP | 13.96 | No | ns | 0.0788 | B | WT 1 uM MPEP |
| WT vs. WT 5 μM MPEP | 31.48 | Yes | **** | <0.0001 | C | WT 5 uM MPEP |
| WT vs. WT 10 μM MPEP | 40.84 | Yes | **** | <0.0001 | D | WT 10 uM MPEP |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. WT 1 μM MPEP | 50.04 | 36.07 | 13.96 | 28 | 14 | 2.222 |
| WT vs. WT 5 μM MPEP | 50.04 | 18.56 | 31.48 | 28 | 9 | 4.28 |
| WT vs. WT 10 μM MPEP | 50.04 | 9.2 | 40.84 | 28 | 15 | 6.649 |
See Figure 3a.
| Table analyzed | risperidone 30 sec lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 6 |
| Kruskal-Wallis statistic | 74.44 |
| Data summary | |
| Number of treatments (columns) | 6 |
| Number of values (total) | 155 |
See Figure 3a.
| Number of families | 1 | |||||
| Number of comparisons per family | 15 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT dmso vs. WT risp | 4.607 | No | ns | >0.9999 | A-B | |
| WT dmso vs. shk3n dmso | 50.65 | Yes | *** | 0.0003 | A-C | |
| WT dmso vs. shk3n risp | 78.7 | Yes | **** | <0.0001 | A-D | |
| WT dmso vs. shk3c dmso | 41.9 | Yes | ** | 0.0059 | A-E | |
| WT dmso vs. shk3c risp | 70.75 | Yes | **** | <0.0001 | A-F | |
| WT risp vs. shk3n dmso | 37.29 | Yes | * | 0.0284 | B-C | |
| WT risp vs. shk3n risp | 74.1 | Yes | **** | <0.0001 | B-D | |
| WT risp vs. shk3c dmso | 66.14 | Yes | **** | <0.0001 | B-E | |
| WT risp vs. shk3c risp | 46.05 | Yes | ** | 0.0022 | B-F | |
| shk3n dmso vs. shk3n risp | 28.05 | No | ns | 0.4389 | C-D | |
| shk3n dmso vs. shk3c dmso | -8.756 | No | ns | >0.9999 | C-E | |
| shk3n dmso vs. shk3c risp | 20.1 | No | ns | >0.9999 | C-F | |
| shk3n risp vs. shk3c dmso | -36.81 | No | ns | 0.0578 | D-E | |
| shk3n risp vs. shk3c risp | -7.955 | No | ns | >0.9999 | D-F | |
| shk3c dmso vs. shk3c risp | 28.85 | No | ns | 0.3522 | E-F | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT dmso vs. WT risp | 112.5 | 107.9 | 4.607 | 31 | 29 | 0.4063 |
| WT dmso vs. shk3n dmso | 112.5 | 61.85 | 50.65 | 31 | 24 | 4.235 |
| WT dmso vs. shk3n risp | 112.5 | 33.8 | 78.7 | 31 | 23 | 6.498 |
| WT dmso vs. shk3c dmso | 112.5 | 70.6 | 41.9 | 31 | 25 | 3.545 |
| WT dmso vs. shk3c risp | 112.5 | 41.75 | 70.75 | 31 | 23 | 5.841 |
| WT risp vs. shk3n dmso | 107.9 | 61.85 | 46.05 | 29 | 24 | 3.792 |
| WT risp vs. shk3n risp | 107.9 | 33.8 | 74.1 | 29 | 23 | 6.027 |
| WT risp vs. shk3c dmso | 107.9 | 70.6 | 37.29 | 29 | 25 | 3.107 |
| WT risp vs. shk3c risp | 107.9 | 41.75 | 66.14 | 29 | 23 | 5.38 |
| shk3n dmso vs. shk3n risp | 61.85 | 33.8 | 28.05 | 24 | 23 | 2.18 |
| shk3n dmso vs. shk3c dmso | 61.85 | 70.6 | -8.756 | 24 | 25 | 0.6954 |
| shk3n dmso vs. shk3c risp | 61.85 | 41.75 | 20.1 | 24 | 23 | 1.562 |
| shk3n risp vs. shk3c dmso | 33.8 | 70.6 | -36.81 | 23 | 25 | 2.89 |
| shk3n risp vs. shk3c risp | 33.8 | 41.75 | -7.955 | 23 | 23 | 0.6114 |
| shk3c dmso vs. shk3c risp | 70.6 | 41.75 | 28.85 | 25 | 23 | 2.266 |
See Figure 3b.
| Table analyzed | risperidone 5 min lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 6 |
| Kruskal-Wallis statistic | 27.87 |
| Data summary | |
| Number of treatments (columns) | 6 |
| Number of values (total) | 155 |
See Figure 3b.
| Number of families | 1 | |||||
| Number of comparisons per family | 15 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT dmso vs. WT risp | 0.9774 | No | ns | >0.9999 | A-B | |
| WT dmso vs. shk3n dmso | 45.1 | Yes | ** | 0.0024 | A-C | |
| WT dmso vs. shk3n risp | 32.82 | No | ns | 0.1012 | A-D | |
| WT dmso vs. shk3c dmso | 39.86 | Yes | * | 0.0112 | A-E | |
| WT dmso vs. shk3c risp | 33.23 | No | ns | 0.0914 | A-F | |
| WT risp vs. shk3n dmso | 44.12 | Yes | ** | 0.0042 | B-C | |
| WT risp vs. shk3n risp | 31.84 | No | ns | 0.1442 | B-D | |
| WT risp vs. shk3c dmso | 38.88 | Yes | * | 0.018 | B-E | |
| WT risp vs. shk3c risp | 32.25 | No | ns | 0.1308 | B-F | |
| shk3n dmso vs. shk3n risp | -12.28 | No | ns | >0.9999 | C-D | |
| shk3n dmso vs. shk3c dmso | -5.241 | No | ns | >0.9999 | C-E | |
| shk3n dmso vs. shk3c risp | -11.87 | No | ns | >0.9999 | C-F | |
| shk3n risp vs. shk3c dmso | 7.042 | No | ns | >0.9999 | D-E | |
| shk3n risp vs. shk3c risp | 0.4091 | No | ns | >0.9999 | D-F | |
| shk3c dmso vs. shk3c risp | -6.633 | No | ns | >0.9999 | E-F | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT dmso vs. WT risp | 98.32 | 97.34 | 0.9774 | 31 | 29 | 0.08619 |
| WT dmso vs. shk3n dmso | 98.32 | 53.22 | 45.1 | 31 | 24 | 3.771 |
| WT dmso vs. shk3n risp | 98.32 | 65.5 | 32.82 | 31 | 23 | 2.709 |
| WT dmso vs. shk3c dmso | 98.32 | 58.46 | 39.86 | 31 | 25 | 3.372 |
| WT dmso vs. shk3c risp | 98.32 | 65.09 | 33.23 | 31 | 23 | 2.743 |
| WT risp vs. shk3n dmso | 97.34 | 53.22 | 44.12 | 29 | 24 | 3.633 |
| WT risp vs. shk3n risp | 97.34 | 65.5 | 31.84 | 29 | 23 | 2.59 |
| WT risp vs. shk3c dmso | 97.34 | 58.46 | 38.88 | 29 | 25 | 3.239 |
| WT risp vs. shk3c risp | 97.34 | 65.09 | 32.25 | 29 | 23 | 2.623 |
| shk3n dmso vs. shk3n risp | 53.22 | 65.5 | -12.28 | 24 | 23 | 0.9544 |
| shk3n dmso vs. shk3c dmso | 53.22 | 58.46 | -5.241 | 24 | 25 | 0.4162 |
| shk3n dmso vs. shk3c risp | 53.22 | 65.09 | -11.87 | 24 | 23 | 0.9226 |
| shk3n risp vs. shk3c dmso | 65.5 | 58.46 | 7.042 | 23 | 25 | 0.5528 |
| shk3n risp vs. shk3c risp | 65.5 | 65.09 | 0.4091 | 23 | 23 | 0.03144 |
| shk3c dmso vs. shk3c risp | 58.46 | 65.09 | -6.633 | 25 | 23 | 0.5207 |
See Figure 4a.
| Table analyzed | LiCL 30 sec lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 6 |
| Kruskal-Wallis statistic | 60.74 |
| Data summary | |
| Number of treatments (columns) | 6 |
| Number of values (total) | 104 |
See Figure 4a.
| Number of families | 1 | |||||
| Number of comparisons per family | 15 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT vs. WT LiCL | -5.725 | No | ns | >0.9999 | A-B | |
| WT vs. shk3n hom | 46.9 | Yes | **** | <0.0001 | A-C | |
| WT vs. shk3n hom LiCL | 42.23 | Yes | *** | 0.0002 | A-D | |
| WT vs. shk3c hom | 49.59 | Yes | **** | <0.0001 | A-E | |
| WT vs. shk3c hom LiCL | 43.71 | Yes | *** | 0.0002 | A-F | |
| WT LiCL vs. shk3n hom | 52.63 | Yes | **** | <0.0001 | B-C | |
| WT LiCL vs. shk3n hom LiCL | 47.96 | Yes | **** | <0.0001 | B-D | |
| WT LiCL vs. shk3c hom | 55.31 | Yes | **** | <0.0001 | B-E | |
| WT LiCL vs. shk3c hom LiCL | 49.44 | Yes | **** | <0.0001 | B-F | |
| shk3n hom vs. shk3n hom LiCL | -4.667 | No | ns | >0.9999 | C-D | |
| shk3n hom vs. shk3c hom | 2.688 | No | ns | >0.9999 | C-E | |
| shk3n hom vs. shk3c hom LiCL | -3.188 | No | ns | >0.9999 | C-F | |
| shk3n hom LiCL vs. shk3c hom | 7.354 | No | ns | >0.9999 | D-E | |
| shk3n hom LiCL vs. shk3c hom LiCL | 1.479 | No | ns | >0.9999 | D-F | |
| shk3c hom vs. shk3c hom LiCL | -5.875 | No | ns | >0.9999 | E-F | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. WT LiCL | 81.4 | 87.13 | -5.725 | 20 | 16 | 0.5658 |
| WT vs. shk3n hom | 81.4 | 34.5 | 46.9 | 20 | 18 | 4.785 |
| WT vs. shk3n hom LiCL | 81.4 | 39.17 | 42.23 | 20 | 18 | 4.309 |
| WT vs. shk3c hom | 81.4 | 31.81 | 49.59 | 20 | 16 | 4.901 |
| WT vs. shk3c hom LiCL | 81.4 | 37.69 | 43.71 | 20 | 16 | 4.32 |
| WT LiCL vs. shk3n hom | 87.13 | 34.5 | 52.63 | 16 | 18 | 5.077 |
| WT LiCL vs. shk3n hom LiCL | 87.13 | 39.17 | 47.96 | 16 | 18 | 4.627 |
| WT LiCL vs. shk3c hom | 87.13 | 31.81 | 55.31 | 16 | 16 | 5.186 |
| WT LiCL vs. shk3c hom LiCL | 87.13 | 37.69 | 49.44 | 16 | 16 | 4.635 |
| shk3n hom vs. shk3n hom LiCL | 34.5 | 39.17 | -4.667 | 18 | 18 | 0.4641 |
| shk3n hom vs. shk3c hom | 34.5 | 31.81 | 2.688 | 18 | 16 | 0.2593 |
| shk3n hom vs. shk3c hom LiCL | 34.5 | 37.69 | -3.188 | 18 | 16 | 0.3075 |
| shk3n hom LiCL vs. shk3c hom | 39.17 | 31.81 | 7.354 | 18 | 16 | 0.7095 |
| shk3n hom LiCL vs. shk3c hom LiCL | 39.17 | 37.69 | 1.479 | 18 | 16 | 0.1427 |
| shk3c hom vs. shk3c hom LiCL | 31.81 | 37.69 | -5.875 | 16 | 16 | 0.5508 |
See Figure 5b.
| Table analyzed | LiCL 5 min lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 6 |
| Kruskal-Wallis statistic | 49.22 |
| Data summary | |
| Number of treatments (columns) | 6 |
| Number of values (total) | 104 |
See Figure 5b.
| Number of families | 1 | |||||
| Number of comparisons per family | 15 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT vs. WT LiCL | 9.069 | No | ns | >0.9999 | A-B | |
| WT vs. shk3n hom | 55.96 | Yes | **** | <0.0001 | A-C | |
| WT vs. shk3n hom LiCL | 46.02 | Yes | **** | <0.0001 | A-D | |
| WT vs. shk3c hom | 39.07 | Yes | ** | 0.0017 | A-E | |
| WT vs. shk3c hom LiCL | 40.91 | Yes | *** | 0.0008 | A-F | |
| WT LiCL vs. shk3n hom | 46.89 | Yes | **** | <0.0001 | B-C | |
| WT LiCL vs. shk3n hom LiCL | 36.95 | Yes | ** | 0.0055 | B-D | |
| WT LiCL vs. shk3c hom | 30 | No | ns | 0.0737 | B-E | |
| WT LiCL vs. shk3c hom LiCL | 31.84 | Yes | * | 0.0424 | B-F | |
| shk3n hom vs. shk3n hom LiCL | -9.944 | No | ns | >0.9999 | C-D | |
| shk3n hom vs. shk3c hom | -16.89 | No | ns | >0.9999 | C-E | |
| shk3n hom vs. shk3c hom LiCL | -15.05 | No | ns | >0.9999 | C-F | |
| shk3n hom LiCL vs. shk3c hom | -6.948 | No | ns | >0.9999 | D-E | |
| shk3n hom LiCL vs. shk3c hom LiCL | -5.104 | No | ns | >0.9999 | D-F | |
| shk3c hom vs. shk3c hom LiCL | 1.844 | No | ns | >0.9999 | E-F | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. WT LiCL | 83.85 | 74.78 | 9.069 | 20 | 16 | 0.8963 |
| WT vs. shk3n hom | 83.85 | 27.89 | 55.96 | 20 | 18 | 5.71 |
| WT vs. shk3n hom LiCL | 83.85 | 37.83 | 46.02 | 20 | 18 | 4.695 |
| WT vs. shk3c hom | 83.85 | 44.78 | 39.07 | 20 | 16 | 3.861 |
| WT vs. shk3c hom LiCL | 83.85 | 42.94 | 40.91 | 20 | 16 | 4.044 |
| WT LiCL vs. shk3n hom | 74.78 | 27.89 | 46.89 | 16 | 18 | 4.524 |
| WT LiCL vs. shk3n hom LiCL | 74.78 | 37.83 | 36.95 | 16 | 18 | 3.565 |
| WT LiCL vs. shk3c hom | 74.78 | 44.78 | 30 | 16 | 16 | 2.813 |
| WT LiCL vs. shk3c hom LiCL | 74.78 | 42.94 | 31.84 | 16 | 16 | 2.986 |
| shk3n hom vs. shk3n hom LiCL | 27.89 | 37.83 | -9.944 | 18 | 18 | 0.989 |
| shk3n hom vs. shk3c hom | 27.89 | 44.78 | -16.89 | 18 | 16 | 1.63 |
| shk3n hom vs. shk3c hom LiCL | 27.89 | 42.94 | -15.05 | 18 | 16 | 1.452 |
| shk3n hom LiCL vs. shk3c hom | 37.83 | 44.78 | -6.948 | 18 | 16 | 0.6703 |
| shk3n hom LiCL vs. shk3c hom LiCL | 37.83 | 42.94 | -5.104 | 18 | 16 | 0.4924 |
| shk3c hom vs. shk3c hom LiCL | 44.78 | 42.94 | 1.844 | 16 | 16 | 0.1729 |
See Figure 5a.
| Table analyzed | CBZ 30 sec lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 6 |
| Kruskal-Wallis statistic | 73.6 |
| Data summary | |
| Number of treatments (columns) | 6 |
| Number of values (total) | 176 |
See Figure 5a.
| Number of families | 1 | |||||
| Number of comparisons per family | 15 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT vs. WT CBZ | 14.04 | No | ns | >0.9999 | A-B | |
| WT vs. shk3n | 61.73 | Yes | **** | <0.0001 | A-C | |
| WT vs. shk3n CBZ | 112.4 | Yes | **** | <0.0001 | A-D | |
| WT vs. shk3c | 42.38 | Yes | * | 0.0175 | A-E | |
| WT vs. shk3c CBZ | 39.99 | Yes | * | 0.0167 | A-F | |
| WT CBZ vs. shk3n | 47.69 | Yes | ** | 0.0053 | B-C | |
| WT CBZ vs. shk3n CBZ | 98.35 | Yes | **** | <0.0001 | B-D | |
| WT CBZ vs. shk3c | 28.34 | No | ns | 0.4199 | B-E | |
| WT CBZ vs. shk3c CBZ | 25.95 | No | ns | 0.4803 | B-F | |
| shk3n vs. shk3n CBZ | 50.66 | Yes | * | 0.0181 | C-D | |
| shk3n vs. shk3c | -19.35 | No | ns | >0.9999 | C-E | |
| shk3n vs. shk3c CBZ | -21.74 | No | ns | >0.9999 | C-F | |
| shk3n CBZ vs. shk3c | -70.01 | Yes | **** | <0.0001 | D-E | |
| shk3n CBZ vs. shk3c CBZ | -72.39 | Yes | **** | <0.0001 | D-F | |
| shk3c vs. shk3c CBZ | -2.383 | No | ns | >0.9999 | E-F | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. WT CBZ | 126.2 | 112.2 | 14.04 | 35 | 37 | 1.169 |
| WT vs. shk3n | 126.2 | 64.5 | 61.73 | 35 | 24 | 4.571 |
| WT vs. shk3n CBZ | 126.2 | 13.84 | 112.4 | 35 | 19 | 7.741 |
| WT vs. shk3c | 126.2 | 83.85 | 42.38 | 35 | 27 | 3.247 |
| WT vs. shk3c CBZ | 126.2 | 86.24 | 39.99 | 35 | 34 | 3.26 |
| WT CBZ vs. shk3n | 112.2 | 64.5 | 47.69 | 37 | 24 | 3.571 |
| WT CBZ vs. shk3n CBZ | 112.2 | 13.84 | 98.35 | 37 | 19 | 6.839 |
| WT CBZ vs. shk3c | 112.2 | 83.85 | 28.34 | 37 | 27 | 2.197 |
| WT CBZ vs. shk3c CBZ | 112.2 | 86.24 | 25.95 | 37 | 34 | 2.144 |
| shk3n vs. shk3n CBZ | 64.5 | 13.84 | 50.66 | 24 | 19 | 3.238 |
| shk3n vs. shk3c | 64.5 | 83.85 | -19.35 | 24 | 27 | 1.354 |
| shk3n vs. shk3c CBZ | 64.5 | 86.24 | -21.74 | 24 | 34 | 1.6 |
| shk3n CBZ vs. shk3c | 13.84 | 83.85 | -70.01 | 19 | 27 | 4.589 |
| shk3n CBZ vs. shk3c CBZ | 13.84 | 86.24 | -72.39 | 19 | 34 | 4.96 |
| shk3c vs. shk3c CBZ | 83.85 | 86.24 | -2.383 | 27 | 34 | 0.1815 |
See Figure 5b.
| Table analyzed | CBZ 5 min lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 6 |
| Kruskal-Wallis statistic | 40.2 |
| Data summary | |
| Number of treatments (columns) | 6 |
| Number of values (total) | 176 |
See Figure 5b.
| Number of families | 1 | |||||
| Number of comparisons per family | 15 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT vs. WT cbz | 25.72 | No | ns | 0.4844 | A-B | |
| WT vs. shank3n | 62.12 | Yes | *** | 0.0003 | A-C | |
| WT vs. shank3n CBZ | 72.55 | Yes | **** | <0.0001 | A-D | |
| WT vs. shank3c | 51.91 | Yes | ** | 0.001 | A-E | |
| WT vs. shank3c CBZ | 48.3 | Yes | ** | 0.0012 | A-F | |
| WT cbz vs. shank3n | 36.4 | No | ns | 0.1706 | B-C | |
| WT cbz vs. shank3n CBZ | 46.83 | Yes | ** | 0.0068 | B-D | |
| WT cbz vs. shank3c | 26.19 | No | ns | 0.6338 | B-E | |
| WT cbz vs. shank3c CBZ | 22.58 | No | ns | 0.9316 | B-F | |
| shank3n vs. shank3n CBZ | 10.43 | No | ns | >0.9999 | C-D | |
| shank3n vs. shank3c | -10.2 | No | ns | >0.9999 | C-E | |
| shank3n vs. shank3c CBZ | -13.82 | No | ns | >0.9999 | C-F | |
| shank3n CBZ vs. shank3c | -20.64 | No | ns | >0.9999 | D-E | |
| shank3n CBZ vs. shank3c CBZ | -24.25 | No | ns | >0.9999 | D-F | |
| shank3c vs. shank3c CBZ | -3.611 | No | ns | >0.9999 | E-F | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT vs. WT cbz | 127.8 | 102.1 | 25.72 | 35 | 37 | 2.141 |
| WT vs. shank3n | 127.8 | 65.68 | 62.12 | 35 | 19 | 4.278 |
| WT vs. shank3n CBZ | 127.8 | 55.25 | 72.55 | 35 | 24 | 5.373 |
| WT vs. shank3c | 127.8 | 75.89 | 51.91 | 35 | 27 | 3.978 |
| WT vs. shank3c CBZ | 127.8 | 79.5 | 48.3 | 35 | 34 | 3.937 |
| WT cbz vs. shank3n | 102.1 | 65.68 | 36.4 | 37 | 19 | 2.531 |
| WT cbz vs. shank3n CBZ | 102.1 | 55.25 | 46.83 | 37 | 24 | 3.507 |
| WT cbz vs. shank3c | 102.1 | 75.89 | 26.19 | 37 | 27 | 2.031 |
| WT cbz vs. shank3c CBZ | 102.1 | 79.5 | 22.58 | 37 | 34 | 1.866 |
| shank3n vs. shank3n CBZ | 65.68 | 55.25 | 10.43 | 19 | 24 | 0.6669 |
| shank3n vs. shank3c | 65.68 | 75.89 | -10.2 | 19 | 27 | 0.6688 |
| shank3n vs. shank3c CBZ | 65.68 | 79.5 | -13.82 | 19 | 34 | 0.9467 |
| shank3n CBZ vs. shank3c | 55.25 | 75.89 | -20.64 | 24 | 27 | 1.444 |
| shank3n CBZ vs. shank3c CBZ | 55.25 | 79.5 | -24.25 | 24 | 34 | 1.785 |
| shank3c vs. shank3c CBZ | 75.89 | 79.5 | -3.611 | 27 | 34 | 0.2749 |
See Figure 6a.
| Table analyzed | MPEP 30 sec lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.001 |
| Exact or approximate P value? | Approximate |
| P value summary | *** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 6 |
| Kruskal-Wallis statistic | 45.04 |
| Data summary | |
| Number of treatments (columns) | 6 |
| Number of values (total) | 153 |
See Figure 6a.
| Number of families | 1 | |||||
| Number of comparisons per family | 15 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| shank3+/+ DMSO vs. shank3+/+ MPEP | 48.38 | Yes | ** | 0.004 | A-B | |
| shank3+/+ DMSO vs. shank3n DMSO | 57.11 | Yes | *** | <0.001 | A-C | |
| shank3+/+ DMSO vs. shank3n MPEP | 65.55 | Yes | *** | <0.001 | A-D | |
| shank3+/+ DMSO vs. shank3c DMSO | 53.83 | Yes | ** | 0.001 | A-E | |
| shank3+/+ DMSO vs. shank3c MPEP | 71.91 | Yes | *** | <0.001 | A-F | |
| shank3+/+ MPEP vs. shank3n DMSO | 8.724 | No | ns | >0.99 | B-C | |
| shank3+/+ MPEP vs. shank3n MPEP | 17.17 | No | ns | >0.99 | B-D | |
| shank3+/+ MPEP vs. shank3c DMSO | 5.45 | No | ns | >0.99 | B-E | |
| shank3+/+ MPEP vs. shank3c MPEP | 23.53 | No | ns | >0.99 | B-F | |
| shank3n DMSO vs. shank3n MPEP | 8.445 | No | ns | >0.99 | C-D | |
| shank3n DMSO vs. shank3c DMSO | -3.274 | No | ns | >0.99 | C-E | |
| shank3n DMSO vs. shank3c MPEP | 14.8 | No | ns | >0.99 | C-F | |
| shank3n MPEP vs. shank3c DMSO | -11.72 | No | ns | >0.99 | D-E | |
| shank3n MPEP vs. shank3c MPEP | 6.358 | No | ns | >0.99 | D-F | |
| shank3c DMSO vs. shank3c MPEP | 18.08 | No | ns | >0.99 | E-F | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| shank3+/+ DMSO vs. shank3+/+ MPEP | 127.3 | 78.95 | 48.38 | 23 | 24 | 3.677 |
| shank3+/+ DMSO vs. shank3n DMSO | 127.3 | 70.23 | 57.11 | 23 | 33 | 4.864 |
| shank3+/+ DMSO vs. shank3n MPEP | 127.3 | 61.78 | 65.55 | 23 | 31 | 5.624 |
| shank3+/+ DMSO vs. shank3c DMSO | 127.3 | 73.5 | 53.83 | 23 | 19 | 3.967 |
| shank3+/+ DMSO vs. shank3c MPEP | 127.3 | 55.42 | 71.91 | 23 | 23 | 5.868 |
| shank3+/+ MPEP vs. shank3n DMSO | 78.95 | 70.23 | 8.724 | 24 | 33 | 0.682 |
| shank3+/+ MPEP vs. shank3n MPEP | 78.95 | 61.78 | 17.17 | 24 | 31 | 1.35 |
| shank3+/+ MPEP vs. shank3c DMSO | 78.95 | 73.5 | 5.45 | 24 | 19 | 0.3761 |
| shank3+/+ MPEP vs. shank3c MPEP | 78.95 | 55.42 | 23.53 | 24 | 23 | 1.774 |
| shank3n DMSO vs. shank3n MPEP | 70.23 | 61.78 | 8.445 | 33 | 30 | 0.7513 |
| shank3n DMSO vs. shank3c DMSO | 70.23 | 73.5 | -3.274 | 33 | 19 | 0.2477 |
| shank3n DMSO vs. shank3c MPEP | 70.23 | 55.42 | 14.8 | 33 | 23 | 1.248 |
| shank3n MPEP vs. shank3c DMSO | 61.78 | 73.5 | -11.72 | 31 | 19 | 0.8918 |
| shank3n MPEP vs. shank3c MPEP | 61.78 | 55.42 | 6.358 | 31 | 23 | 0.5399 |
| shank3c DMSO vs. shank3c MPEP | 73.5 | 55.42 | 18.08 | 19 | 23 | 1.322 |
See Figure 6b.
| Table analyzed | shank3 MPEP 5 min lights-off |
|---|---|
| Kruskal-Wallis test | |
| P value | <0.0001 |
| Exact or approximate P value? | Approximate |
| P value summary | **** |
| Do the medians vary signif. (P < 0.05)? | Yes |
| Number of groups | 6 |
| Kruskal-Wallis statistic | 27 |
| Data summary | |
| Number of treatments (columns) | 6 |
| Number of values (total) | 153 |
See Figure 6b.
| Number of families | 1 | |||||
| Number of comparisons per family | 15 | |||||
| Alpha | 0.05 | |||||
| Dunn's multiple comparisons test | Mean rank diff. | Significant? | Summary | Adjusted P Value | ||
| WT dmso vs. WT mpep | 40.31 | Yes | * | 0.0225 | A-B | |
| WT dmso vs. shk3n dmso | 50.63 | Yes | *** | 0.0003 | A-C | |
| WT dmso vs. shk3 mpep | 49.3 | Yes | *** | 0.0006 | A-D | |
| WT dmso vs. shk3c dmso | 39.96 | Yes | * | 0.0473 | A-E | |
| WT dmso vs. shk3c mpep | 58.7 | Yes | **** | <0.0001 | A-F | |
| WT mpep vs. shk3n dmso | 10.32 | No | ns | >0.9999 | B-C | |
| WT mpep vs. shk3 mpep | 8.994 | No | ns | >0.9999 | B-D | |
| WT mpep vs. shk3c dmso | -0.3502 | No | ns | >0.9999 | B-E | |
| WT mpep vs. shk3c mpep | 18.4 | No | ns | >0.9999 | B-F | |
| shk3n dmso vs. shk3 mpep | -1.329 | No | ns | >0.9999 | C-D | |
| shk3n dmso vs. shk3c dmso | -10.67 | No | ns | >0.9999 | C-E | |
| shk3n dmso vs. shk3c mpep | 8.074 | No | ns | >0.9999 | C-F | |
| shk3 mpep vs. shk3c dmso | -9.344 | No | ns | >0.9999 | D-E | |
| shk3 mpep vs. shk3c mpep | 9.403 | No | ns | >0.9999 | D-F | |
| shk3c dmso vs. shk3c mpep | 18.75 | No | ns | >0.9999 | E-F | |
| Test details | Mean rank 1 | Mean rank 2 | Mean rank diff. | n1 | n2 | Z |
| WT dmso vs. WT mpep | 115.1 | 74.76 | 40.31 | 23 | 24 | 3.174 |
| WT dmso vs. shk3n dmso | 115.1 | 64.44 | 50.63 | 23 | 33 | 4.293 |
| WT dmso vs. shk3 mpep | 115.1 | 65.77 | 49.3 | 23 | 31 | 4.125 |
| WT dmso vs. shk3c dmso | 115.1 | 75.11 | 39.96 | 23 | 19 | 2.953 |
| WT dmso vs. shk3c mpep | 115.1 | 56.36 | 58.7 | 23 | 23 | 4.573 |
| WT mpep vs. shk3n dmso | 74.76 | 64.44 | 10.32 | 24 | 33 | 0.8869 |
| WT mpep vs. shk3 mpep | 74.76 | 65.77 | 8.994 | 24 | 31 | 0.7622 |
| WT mpep vs. shk3c dmso | 74.76 | 75.11 | -0.3502 | 24 | 19 | 0.02614 |
| WT mpep vs. shk3c mpep | 74.76 | 56.36 | 18.4 | 24 | 23 | 1.449 |
| shk3n dmso vs. shk3 mpep | 64.44 | 65.77 | -1.329 | 33 | 30 | 0.1228 |
| shk3n dmso vs. shk3c dmso | 64.44 | 75.11 | -10.67 | 33 | 19 | 0.8508 |
| shk3n dmso vs. shk3c mpep | 64.44 | 56.36 | 8.074 | 33 | 23 | 0.6847 |
| shk3 mpep vs. shk3c dmso | 65.77 | 75.11 | -9.344 | 31 | 19 | 0.7361 |
| shk3 mpep vs. shk3c mpep | 65.77 | 56.36 | 9.403 | 31 | 23 | 0.7868 |
| shk3c dmso vs. shk3c mpep | 75.11 | 56.36 | 18.75 | 19 | 23 | 1.385 |
See Figure 7b.
See Figure 7b.
See Figure 7b.
See Figure 7b.
DRYAD: Drugs prescribed for Phelan-McDermid syndrome differentially impact sensory behaviors in shank3 zebrafish models. https://doi.org/10.5061/dryad.hqbzkh1kn (Kozol & Dallman, 2023).
DRYAD: ARRIVE checklist for ‘Drugs prescribed for Phelan-McDermid syndrome differentially impact sensory behaviors in shank3 zebrafish models.’ https://doi.org/10.5061/dryad.hqbzkh1kn (Kozol & Dallman, 2023).
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
We would like to thank the Phelan-McDermid Syndrome Foundation for including us in their scientific/family conferences where we learned about the challenges families face in managing multiple symptoms. We also thank Ricardo Cepeda for his excellent care of the zebrafish.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Phelan-McDermid syndrome
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 27 Sep 23 |
read | read |
|
Version 1 23 Jan 23 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)