Keywords
Dexistes rikuzenius, Pleuronectiformes, Mitogenome
This article is included in the Genomics and Genetics gateway.
Dexistes rikuzenius, Pleuronectiformes, Mitogenome
The family Pleuronectidae belongs to the order Pleuronectiformes. There are 25 genera and 62 species in the family as listed in FishBase (Froese and Pauly, 2022), while Eschmeyer’s Catalog of Fishes (Fricke et al., 2022) reports 24 genera and 64 species. Further different classifications in the literature assume 23 genera and 61 species (Nelson et al., 2016), or 24 genera and 59 species (Vinnikov et al., 2018) based on genetic analysis. However, only 25 species in 18 genera have been reported to occur in Korea (MABIK, 2017).
The Rikuzen flounder (Dexistes rikuzenius), which is placed in this family, is a flatfish that lives on sandy or gravelly bottoms at depths of 42-200 meters (Yamada et al., 1995) and is distributed in the South Sea of South Korea, the East China Sea, and southern Hokkaido of Japan. The Rikuzen flounder is one of the rarest species in the family Pleuronectidae in South Korea. Its larvae are reported to be morphologically very similar to those of the stone flounder (Platichthys bicoloratus) (Lee et al., 2019), but little morphological and genetic information on this species is available. The mitogenomic data presented here are therefore expected to provide information that will aid in the efficient resource management of this species and further the molecular phylogenetic taxonomy of the family Pleuronectidae.
The specimen used in this study was secured from Jeju South Korea (33°571’N, 126°256’E) and the storage facility of the Marine Fish Resource Bank of Korea in Pukyong National University were used to deposit under Voucher no. PKU 56422 (Jinkoo Kim, tjgk2002@gmail.com).
The genomic DNA (gDNA) was extracted from a fin of the specimen using the PureHelixTM Genomic DNA Prep Kit [Animal], Solution Type (NANOHELIX, Daejeon, South Korea). PCR was conducted using the fish universal primer set (Ward et al., 2005) to amplify the cox1 gene and the nucleotide sequence was analyzed by Macrogen (Daejeon, South Korea). A BLASTN (Johnson et al., 2008) search were utilized to confirm the species.
gDNA (1 μg) was sheared using the S220 Ultra sonicator (Covaris, Woburn, USA). MGIEasy DNA library prep kit (MGI Tech, Shenzhen, China) was used for library preparation with according to the manufacturer’s instructions. Briefly, Fragmented gDNA was selected based on its size using AMPure XP magnetic beads and the fragmented gDNA was end-repaired and a-tailed at 37 °C for 30 min, and 65 °C for 15 min. Indexing adapter was ligated to the ends of the DNA fragments at 23 °C for 60 min. PCR was performed to enrich those DNA fragments that have adapter molecules after purifying the adapter-ligated DNA. Thermocycler conditions were as follows: 95°C for 3 min, 7 cycles of 98°C for 20 s, 60°C for 15 s, and 72°C for 30 s, with a final extension at 72°C for 10 min. The double stranded library is quantified using QauntiFluor ONE dsDNA System (Promega, Madison, USA). The library is circularized at 37 °C for 30 min, and then digested at 37 °C for 30 min, followed by cleanup of circularization product. The library is incubated at 30 °C for 25 min using DNB enzyme for making DNA nanoball (DNB). Finally, Library was quantified by QauntiFluor ssDNA System (Promega, Madison, USA). Sequencing of the prepared DNB was conducted on an MGISEQ-2000 sequencing platform (MGI Tech, Shenzhen, China) with 150 bp paired-end reads.
The raw sequencing data were deposited in the Sequence Read Archive (SRA) database (SRR21644379) (theMOAGEN, 2022b). Trimming was conducted using Cutadapt ver. 4.1 (Martin, 2011), and these trimmed data were assembled using the default option in the de novo assembler of the CLC Genomics Workbench (ver. 20.04; QIAGEN, Venlo, Netherlands). The circular form of the mitogenome obtained from the contig sequences was verified by mapping the filtered data onto the contig sequence using the “Map to Reference” tool in the Geneious software (ver. 2021.2.2; https://www.geneious.com). This final sequence was annotated using the MITOS Webserver (Bernt et al., 2013), and SnapGene software (ver. 5.3.2; GSL Biotech LLC; snapgene.com) was utilized for manually correcting the detailed annotation. Finally, the completed circular form of the mitogenome sequence was registered at the NCBI GenBank (OP066371). A maximum likelihood (ML) phylogenetic tree was constructed with MEGA 11 software (Tamura et al., 2021) using GenBank mitogenomes from all available 18 species of the family Pleuronectidae and one member of the family Bothidae, Bothus myriaster (Accession No. KJ433563), as an outgroup. All mitogenome sequences were collected from GenBank. The nucleotide sequences of PCGs were gathered from each mitogenome sequence and were aligned using the ClustalW multiple alignment tool in BioEdit under default options. The phylogenetic analysis was conducted using the GTR + G + I model with 1,000 bootstrap replicates.
The complete mitochondrial genome of the D. rikuzenius was a circular molecule, 17,494 bp in length (GenBank acc. no. OP066371) (Chae and Kim, 2022), and contained 13 PCGs, 22 tRNA genes, two rRNA genes, and one control region. Proportional nucleotide composition of the mitochondrial genome was A: 28.80% (5,039 bp), T: 26.51% (4,638 bp), C: 28.72% (5,025 bp) and G: 15.96% (2,792 bp), with a combined A+T content of 55.32%.
Among the 37 detected mitochondrial genes, nine of the PCGs and eight tRNA genes (tRNAAla, tRNAAsn, tRNACys, tRNAGln, tRNAGlu, tRNAPro, tRNASer and tRNATyr) were encoded on the L-strand, while all other genes were encoded on the H-strand. Most mitochondrial protein-coding genes had ‘ATG’ as their start codon, whereas only COX1 started with ‘GTG’. The seven PCGs (ATP6, ATP8, COX1, ND1, ND4L, ND5 and ND6) used ‘TAA’ as the complete stop codons, whereas COX3 used ‘TA’, and COX2, CYTB, ND2, ND3 and ND4 used ‘T’ as an incomplete stop codon. The latter two instances are presumably completed to ‘TAA’ by posttranscriptional polyadenylation (Ojala et al., 1981). The 12S and 16S rRNA genes were 949 bp and 1,714 bp long, respectively. The 22 tRNA genes ranged in length from 63 bp (tRNACys) to 74 bp (tRNALeu).
The results of the phylogenetic analysis show that D. rikuzenius is closely related to Cleisthenes herzensteini, and that these two are sister taxa to the two genera Limanda and Hippoglossoides (Figure 1). This is the only study to report a complete mitochondrial DNA sequence in the genus Dexistes. Mitochondrial DNA is a powerful tool for studying the evolution of genomes and for the identification of individual species. Our results will therefore be useful for understanding the phylogenetic relationships and taxonomic classification of the family Pleuronectidae, and are expected to contribute to the efficient resource management of the species.
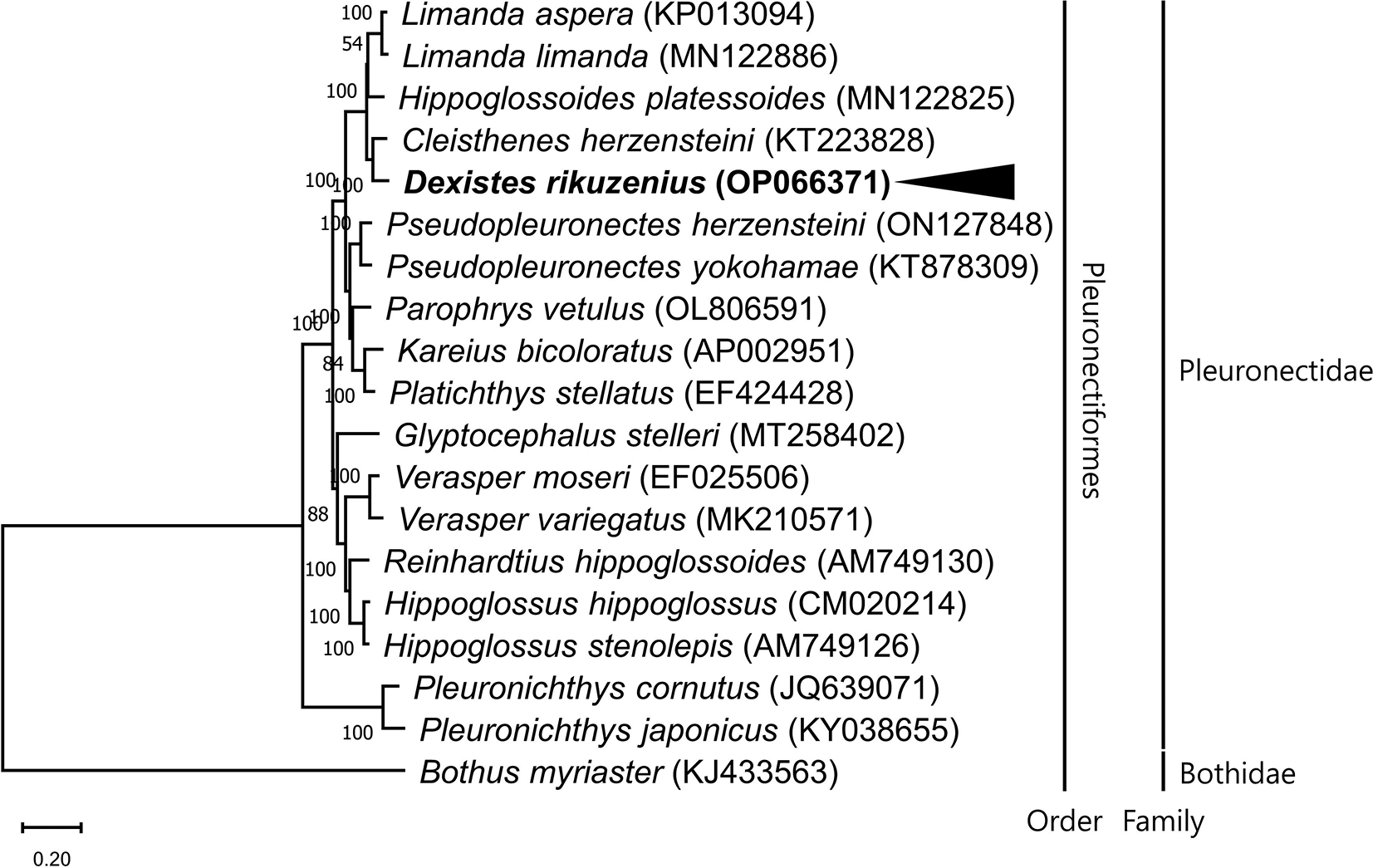
A maximum likelihood dendrogram was constructed using utilizing 13 PCGs from mitogenomes of 18 species in the family Pleuronectidae, and one species in the family Bothidae, Bothus myriaster (Accession No. KJ433563), as an outgroup. The GenBank accession numbers of mitogenome sequences are given in parentheses next to the species names. Node numbers correspond to Bayesian inference's posterior probabilities. The species of interest (Dexistes rikuzenius) is highlighted in bold and with an arrowhead.
No ethical approval is required for this study. We used a flatfish fin from a specimen that was previously collected by the MBRIS, outside of this study. This specimen was dead, and the sample was provided with permission by the MBRIS (permission no. 2022-130).
GenBank: Dexistes rikuzenius mitochondrion, complete genome. Accession number OP066371; https://www.ncbi.nlm.nih.gov/nuccore/OP066371.1 (Chae and Kim, 2022).
BioProject: Dexistes rikuzenius. Accession number PRJNA882492; https://www.ncbi.nlm.nih.gov/bioproject/882492 (theMOAGEN, 2022a).
SRA: Dexistes rikuzenius. Accession number SRR21644379; https://www.ncbi.nlm.nih.gov/sra/SRR21644379 (theMOAGEN, 2022b).
BioSample: Animal sample from Dexistes rikuzenius. Accession number SAMN30941287; https://www.ncbi.nlm.nih.gov/biosample/SAMN30941287 (theMOAGEN 2022c).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Phylogenetic systematics, ichthyology, molecular ecology
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: genomics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 07 Aug 23 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)