Keywords
Echinococcus granulosus, EgFABP1, fatty acids, gene ontology, quantum mechanics, molecular dynamics simulation, MM/GBSA
The zoonotic infection caused by tapeworms Echinococcus is a neglected tropical disease in poor regions with limited access to suitable sanitary conditions. Hydatid cysts produced by Echinococcus granulosus use fatty-acid-binding proteins (FABP) to obtain the fatty acids and cholesterol necessary for their survival from the host. In this work, we analyzed the behaviour of saturated, monounsaturated, and polyunsaturated fatty acids against EgFABP1.
We used computational biology and chemistry techniques and binding free energy estimations by molecular mechanics generalized Born surface area (MM/GBSA).
This research has enabled us to clarify the EgFABP1 isoforms identified in the database, suggesting their potential involvement in diverse cellular activities of Echinococcus granulosus. Conversely, examining the global and local chemical reactivity of 14 fatty acids revealed that liposolubility is contingent upon the degree of unsaturation in the FAs. Additionally, FAs exhibited acceptable levels of oral absorption and bioavailability. The binding of EgFABP1 with FAs analyzed by molecular dynamics simulation showed us that these are highly stable, where the best affinity was with docosahexaenoic acid.
Our results suggest that the action of fatty acids could play an interesting role in detecting early Echinococcus granulosus.
Echinococcus granulosus, EgFABP1, fatty acids, gene ontology, quantum mechanics, molecular dynamics simulation, MM/GBSA
List of Abbreviations – Reorganized
Introduction – Added information related to Platyhelminthes and EgFABP1 solubility at the suggestion of a reviewer.
Computational Details – Molecular geometry optimization. Added experimental references at the suggestion of a reviewer.
Computational Details – Docking determination and molecular dynamics simulation of complex. Added the 25 ns time, which was not included in v1.
Results and Discussion – Computational biology analysis. Added a more detailed description of Figure 1A, at the recommendation of one of the reviewers.
Results and Discussion – Docking and molecular dynamics simulation. Added the first paragraph on the validation of the crystal protein used in this work, based on the recommendation of one of the reviewers.
Results and Discussion – Docking and molecular dynamics simulation. Table 5 on RMSD, RMSF, RG, and SASA from the first version was placed in the Supporting Information as Table S3. Therefore, the table numbering had to be reorganized.
Results and Discussion – Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) estimation. VAC was changed to TVA, due to a typographical error.
Results and Discussion – Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) estimation. A paragraph was added with information on other parasite species that were analyzed with DHA.
Results and Discussion – Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) estimation. A summary paragraph on the action of the proposed FAs against EgFABP1 was added.
Conclusions – The conclusions were improved at the suggestion of the reviewers.
Data availability – The Tables were reorganized.
See the authors' detailed response to the review by Roberto Kopke Salinas
See the authors' detailed response to the review by Guangyou Yang
See the authors' detailed response to the review by Diana G Rios-Valencia
Parasitic nematodes are responsible for significant human suffering, with an astounding 1.5 billion individuals suffering from helminth infections brought on by soil. They infect people, injure cattle, and reduce food production, which results in morbidity and mortality on a global scale.1 Platyhelminthes, specifically cestodes of the genus Echinococcus, are responsible for extremely serious diseases that lead to significant human suffering, mortality, morbidity, and large economic losses, with millions of individuals at risk of infection globally.2 The cestode species of the genus Echinococcus,3–5 currently divided into the species Echinococcus granulosus sensu lato species complex, Echinococcus multilocularis, Echinococcus shiquicus, Echinococcus vogeli, and Echinococcus oligarthra, cause echinococcosis, also known as hydatid disease, and it is estimated that over a million people are afflicted worldwide.6 While the disease manifests clinically as cystic (CE), alveolar (AE), and neotropical (NE) echinococcosis, CE is the most common worldwide, producing substantial morbidity and relative mortality among human populations and is caused by E. granulosus complex.7
Benzimidazoles (BMZ), specifically albendazole and mebendazole when albendazole is not well tolerated, are the only medications now available against CE and AE in clinical settings. In CE, BMZ is additionally used to stop recurrence caused by protoscolece leakage following surgery or percutaneous treatment, either by itself or in combination.8 Much of the parasites’ success is due to their ability to disguise themselves from host immune defenses. However, despite the importance of these organisms to human and animal health, little is understood about the molecular mechanisms that underpin these stealth processes. Increasing our knowledge of how parasitic nematodes influence the immune system has tremendous potential to guide the development of new therapies for immunological dysregulation.9
The acquisition, storage, and transportation of lipids for the formation of hydatid cysts is made possible in this situation by the E. granulosus fatty-acid-binding protein 1 (EgFABP1). To promote the uptake of lipids, the ligand-binding association with EgFABP1 causes a conformational change in the protein structure, improving their intracellular solubility.10 This modification of the interaction mechanism with membranes is also feasible,11 which has made fatty acids (FAs) interesting drug targets. These are classified as saturated fatty acids (SFAs, without double bonds) and unsaturated fatty acids (UFAs, with the presence of double bonds). Also, UFAs are classified as monounsaturated fatty acids (MUFAs) which have only one double bond, and polyunsaturated fatty acids (PUFAs) which have two or more double bonds.12
It has been demonstrated that PUFAs have anti-inflammatory and antibacterial effects, including their effectiveness in protecting against several parasite-related disorders.13 Jae-Won Choi et al.14 suggested that ()-3 PUFAs could be a potential therapeutic candidate for preventing diseases such as toxoplasmosis and other parasitic infections by intracellular protozoa. Moreover, Alhusseiny, S. M. and EI-Beshbishi, S. N.13,15 found that -PUFAs can be considered as potential adjuvant therapeutic agents to anti-parasitic drugs. However, according to Katdare Aakash et al.16 there are many studies of fatty acids as therapeutic additives, but more studies are still required to confirm these as unique therapeutic agents. Because of this, there is a need for greater research to determine how effective certain FAs are at preventing parasite infections caused by E. granulosus.
In this study, we evaluated the binding free energy of 14 fatty acids against EgFABP1 using molecular mechanics generalized Born surface area (MM/GBSA) methodology. Additionally, EgFABP1 analysis was performed regarding phylogeny and gene ontology (GO) enrichment analysis and protein-protein interaction network analysis. The study also delved into the absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of various fatty acids, coupled with an examination of their chemical reactivity using quantum mechanics. Finally, an in-depth exploration was undertaken through all-atom molecular dynamics simulations to analyze the binding free energy of the EgFABP1 complex.
The FASTA sequence of the Echinococcus granulosus fatty acid-binding protein 1 (EgFABP1) protein from Echinococcus granulosus (ID: Q02970) was retrieved from the UniProt database (http://www.uniprot.org/),17 and subjected to the Blastp tool,18 followed by a sequence similarity search performed on E. granulosus (taxid:6210) within the non-redundant protein sequence database. To investigate the evolutionary relationship of EgFABP1 proteins, the retrieved sequences were uploaded in MegaX software and aligned using ClustalW.19 Utilizing the neighbor-joining technique utilized by iTOL, the resulting multiple sequence alignments was used to reconstruct and show a distance-based phylogenetic tree.20 Also, the EgFABP1-related sequences were uploaded into the Cytoscape platform,21 where the plugin StringApp21 was used to retrieve and extend the molecular network information from the STRING database22 and the plugin cytoHubba was used to score and rank the nodes according to network properties, affording to the Maximal Clique Centrality (MCC) topological analysis method.23 The default parameters of Cytoscape were used to show the network; node size and color were manually changed according to the scores obtained from the MCC analysis. Finally, StringApp was also used to retrieve functional enrichment for Gene Ontology (GO) terms, regarding Biological Processes (BP), Molecular Functions (MF), and Cellular Components (CC), while the results were expressed in bubble plots employing the SRplot online platform (http://www.bioinformatics.com.cn/srplot).
In this study, fourteen fatty acids (FAs) were identified from a range of experimental investigations.11,14,24,25 The PubChem CID access code and chemical structure are described in Table 1. The geometries of these compounds were optimized using B3LYP/6-311++g(d,p) basis set level, implemented in ORCA software version 4.1.2.26,27 The ground state was verified by counting the imaginary frequencies of each compound.
Global reactivity descriptors
Global reactivity descriptors are derived from conceptual Density Functional Theory (DFT),28 this type of analysis allows us to understand the relationships among structure, stability and reactivity.29 The global reactivity descriptors were calculated by the mean Koopmans’ theorem, which considers the HOMO (Highest Occupied Molecular Orbital) and LUMO (Lowest Unoccupied Molecular Orbital) energy orbitals (Table 2).
| Reactivity descriptors | Equation | References |
|---|---|---|
| Ionization Potential () | a | 30,31 |
| Electroaffinity () | b | 30,31 |
| Chemical Potential () | 30,32,33 | |
| Global Hardness () | 30,32,33 | |
| Global Softness () | 30,32,33 | |
| Electronegativity () | 30,32,33 | |
| Electrophilicity () | 34,35 | |
| Electron Acceptor () | 36 | |
| Electron Donator () | 35,36 | |
| Net Electrophilicity () | 37 | |
| HOMO-LUMO gap ( gap) |
Local reactivity descriptors: Fukui function
Electrophilic, nucleophilic and radical attacks were calculated using Fukui functions.38 The optimized geometries were selected compounds, and single point energy calculation was performed on the neutral, positive and negatively charged molecule using B3LYP/6-311++g(d,p) basis set implemented in ORCA software version 4.1.2.26,27 The Fukui functions were calculated using Multiwfn39 according to the following equations:
Where is the electron density at a point in space around the molecule. The corresponds to the number of electrons in the molecule. corresponds to an anion with an electron added to the LUMO of the neutral molecule. is the cation with an electron removed from the HOMO of the neutral.
The Molecular Weight (MW), Hydrogen Bonds (nHA, nHB), Polar Surface Area (PSA), Dissociation Constant (pKa), the logarithm of the molar solubility in water (logS), n-octanol/water partition coefficient (logP), n-octanol/buffer solution distribution coefficient at pH = 7.4 (logD7.4), and ADMET pharmacokinetic properties (absorption, distribution, metabolism, excretion, and toxicity) were performed on the ADMETlab 2.0 online platform (https://admetmesh.scbdd.com/). The prediction of pKa values was obtained by MolGpKa web server (https://xundrug.cn/molgpka).
The protein structure of Echinococcus granulosus fatty-acid-binding protein I (EgFABP1) was downloaded from the Protein Data Bank40,41 (https://www.rcsb.org) under access code PDB ID: 1O8V. This protein presents a hydroxylated cysteine (CSO63) that was considered in this work. We defined the AMBER99SB-ILDN force field parameters of the CSO63 and fourteen FAs in the ACPYPE42 server online (https://www.bio2byte.be/login/?next=/acpype/). Finally, the water molecules present in the crystal structure were removed.
The coupling systems were solved in the DockThor server43,44 (https://dockthor.lncc.br/v2) with a grid dimension of 5 × -7 × 10, the genetic algorithm settings with number of evaluations = 1000000, population size = 750, and number of runs = 24.
The molecular dynamics (MD) simulation was carried out using GROMACS v.2020.645 with the AMBER99SB-ILDN force field. Fifteen MD simulations, EgFABP1 without ligand and EgFABP1 coupled to fourteen FAs was considered. The topologies were prepared considering a cubic box of 1 nm from the edge of the box to the protein surface. The TIP3P water model was used for the solvation of the different systems. 200000 energy minimization steps were settled with the steep-descent algorithm. Equilibrium dynamics simulations in the canonical ensemble at constant volume and the production simulation in the isobaric-isothermal at constant pressure (1 bar of reference pressure with Parrinello-Rahman barostat) for a time period of 25 ns, were applied. The calculation time in isobaric-isothermal ensemble (NPT) was 200 ns. In both MD simulations, the V-rescale thermostat regulated the temperature at 309.65 K. The analysis of the MD was carried out with the Gromacs tools and plotted in the Gnuplot v. 4.546 software. Additionally, hydrophobic interactions between FAs and EgFABP1 were analyzed using the PLIP (Protein-Ligand Interaction Profiler) server.47 The graphical visualizations were evaluated in the VMD (Visual Molecular Dynamics)48 and UCSF Chimera v.1.1549 software.
The binding free energy from each coupling system (EgFABP1-FAs) was calculated by Molecular Mechanics-Generalized Born Surface Area (MM/GBSA) method. The mmpbsa.py50 module from AmberTools 2051 program and gmx MMPBSA v1.4.152 were used for these assays. The equations related to calculations of binding free energies () were the following:
Where is the variation between the minimized energy of the protein-ligand complexes (EgFABP1-FAs) that includes the van der Waals () and electrostatic (); is the electrostatic solvation energy, is the difference in surface area energies for protein and ligand and refers to the contribution of entropy to temperature (T).
Spearman’s Rho (also called Spearman’s rank correlation coefficient) was used to estimate the correlation between protein stability and binding free energy of the systems (EgFABP1-FAs). The coefficient was calculated using Python SciPy package.53 Spearman’s Rho is a rank-based non-parametric correlation measurement between two datasets, represented by a monotonic function. The Spearman’s Rho between features X and Y is described by equation 7:
Where denotes the Spearman’s Rho and is the sample size.
According to several studies, E. granulosus comprises a variety of strains with a comparatively high level of genetic variation. Also, numerous aspects of the parasite, such as its life cycle and transmission, pathogenicity, metabolic characteristics, and drug susceptibility, are impacted by this wide range of variety.54 Thirteen proteins with similarity to EgFABP1 were discovered by running a BLASTp search followed by phylogenetic analysis of the amino acid sequences against the E. granulosus proteome. In Figure 1A it is observed that W6UGV8 shares 99% similarity with Q02970, and both are related to W6UFM4, which is a fatty acid-binding protein known as EgFABP1. The phylogenetic analysis indicates that EgFABP1 has a close evolutionary relationship with other fatty acid-binding proteins, including U6JF28, U6JF40, U6JIF2, and W6UDS5. Additionally, EgFABP1 is closely related to other targets such as an immunoglobulin superfamily member (U6J0D8), formin-binding protein (W6UDU7), von Willebrand factor A domain-containing protein 5A (W6UX20), E3 ubiquitin-protein ligase E3D (W6VCE4), and Serine-threonine kinase receptor-associated protein (W6UMK6). Also, understanding how successfully the proteins interfere with the complex regulatory machinery is essential, as developments in network biology have demonstrated that single-protein targeting is ineffective in treating complex disorders55; the analysis of the extended protein network showed that the proteins with the highest centrality scores on the network are annotated as mediators of RNA polymerase II transcription (Figure 1B). Additionally, the analysis of the functional enrichment for Gene Ontology (GO) revealed that the terms linked to BP were retinol metabolic process (GO:0042572) and positive regulation of biological process (GO:0048518), while retinol and fatty acid-binding proteins have been associated to suppress host immunity during nematode infection.56 Also, the GO terms associated with CC found in the analysis were intracellular organelle (GO:0043229) and organelle (GO:0043226), while the MF term was lipid binding (GO:0008289). EgFABP1 isoforms were found in the cytosolic, nuclear, mitochondrial, and microsomal fractions of the experiments, indicating that these molecules may be engaged in a variety of cellular activities57 (Figure 1C).
Global reactivity descriptors
DFT has been very successful in the last decade in predicting the chemical reactivity of various molecules.58–61 Within the DFT theory, one of the most used to determine the stability and reactivity of a particular molecule is the frontier molecular orbitals HOMO and LUMO.62,63 In Table 3, the energy values of the HOMO and LUMO orbitals for all the studied molecules match those reported in the literature for organic molecules.64 It is necessary to highlight that SFAs molecules (MTR, PDN, PLM and STA) showed the most negative values of the HOMO orbital, meaning that these molecules are the most likely to donate electrons in a chemical reaction.63,64 According to the frontier orbital theory, the energy difference between the HOMO and LUMO orbitals (HOMO-LUMO gap) is a measure of the chemical stability of a molecule in a reaction.64,65 High differences between the HOMO concerning the LUMO orbital indicate high stability of the molecule in a chemical reaction.66 Analyzing the HOMO-LUMO gap energy behaviour shown in Table 3, the SFAs molecules obtained the greatest difference in this parameter compared to the other molecules, this means that SFAs compounds present greater chemical stability according to the frontier molecular orbitals theory and the global reactivity descriptors followed by the Koopmans theory,30,59 which means that the HOMO and LUMO frontier orbitals were considered in the calculation.
| Fatty acid | HOMO (eV) | LUMO (eV) | * | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SFAs | MTR | -7.8256 | -0.7020 | 7.1236 | 7.8256 | 0.7020 | -4.2638 | 3.5618 | 0.1404 | 4.2638 | 5.1042 | 5.1292 | 0.8654 | 5.9946 | 4.4949 |
| PDN | -7.8283 | -0.7047 | 7.1236 | 7.8283 | 0.7047 | -4.2665 | 3.5618 | 0.1404 | 4.2665 | 5.1107 | 5.1338 | 0.8673 | 6.0012 | 4.4682 | |
| PLM | -7.8283 | -0.7047 | 7.1236 | 7.8283 | 0.7047 | -4.2665 | 3.5618 | 0.1404 | 4.2665 | 5.1107 | 5.1338 | 0.8673 | 6.0012 | 4.4724 | |
| STA | -7.8283 | -0.7075 | 7.1209 | 7.8283 | 0.7075 | -4.2679 | 3.5604 | 0.1404 | 4.2679 | 5.1159 | 5.1370 | 0.8691 | 6.0060 | 4.4525 | |
| MUFAs | HDN | -6.7154 | -0.3755 | 6.3399 | 6.7154 | 0.3755 | -3.5455 | 3.1700 | 0.1577 | 3.5455 | 3.9654 | 4.1517 | 0.6062 | 4.7579 | 1.7457 |
| OLE | -6.6909 | -0.4245 | 6.2665 | 6.6909 | 0.4245 | -3.5577 | 3.1332 | 0.1596 | 3.5577 | 4.0397 | 4.1904 | 0.6326 | 4.8230 | 1.5998 | |
| PLA | -6.6828 | -0.3728 | 6.3100 | 6.6828 | 0.3728 | -3.5278 | 3.1550 | 0.1585 | 3.5278 | 3.9446 | 4.1306 | 0.6028 | 4.7333 | 1.7051 | |
| TVA | -6.6719 | -0.3537 | 6.3182 | 6.6719 | 0.3537 | -3.5128 | 3.1591 | 0.1583 | 3.5128 | 3.9061 | 4.1044 | 0.5916 | 4.6959 | 1.6521 | |
| CVA | -6.6801 | -0.4218 | 6.2583 | 6.6801 | 0.4218 | -3.5509 | 3.1292 | 0.1598 | 3.5509 | 4.0295 | 4.1813 | 0.6304 | 4.8118 | 1.2909 | |
| PUFAs | ARA | -6.2148 | -0.1170 | 6.0978 | 6.2148 | 0.1170 | -3.1659 | 3.0489 | 0.1640 | 3.1659 | 3.2874 | 3.6077 | 0.4419 | 4.0496 | 2.4058 |
| DHA | -6.3182 | -0.1524 | 6.1658 | 6.3182 | 0.1524 | -3.2353 | 3.0829 | 0.1622 | 3.2353 | 3.3952 | 3.7006 | 0.4653 | 4.1659 | 1.0092 | |
| EPA | -6.3399 | -0.1823 | 6.1576 | 6.3399 | 0.1823 | -3.2611 | 3.0788 | 0.1624 | 3.2611 | 3.4542 | 3.7425 | 0.4814 | 4.2239 | 1.1220 | |
| LIN | -6.7263 | -0.4571 | 6.2692 | 6.7263 | 0.4571 | -3.5917 | 3.1346 | 0.1595 | 3.5917 | 4.1155 | 4.2454 | 0.6537 | 4.8992 | 1.7346 | |
| LNA | -6.2700 | -0.2177 | 6.0523 | 6.2700 | 0.2177 | -3.2438 | 3.0262 | 0.1652 | 3.2438 | 3.4772 | 3.7388 | 0.4949 | 4.2337 | 1.2608 | |
Regarding the dipole moment, we observed that the value decreases depending on the unsaturation degree of the FAs due to their liposolubility.67
To delve into the biological behaviour of the FAs under study, it is necessary to analyze the local reactivity descriptors such as the molecular electrostatic potential and the Fukui function. The molecular electrostatic potential provides information on the atoms of the chemical structure of a molecule with positive or negative charge density; and the Fukui function is a chemical observable that gives us the measure of which part of the molecule is more susceptible to a nucleophilic, electrophilic, or radical attack.
Local reactivity descriptors
As can be seen in Figure 2 ARA, DHA, and EPA molecules (PUFAs with the longest chains) had the most pronounced charge densities of all the compounds studied due to the structure of minimum energy in geometry optimization. These molecules have a greater torsion degree than the others, this implies that their unsaturations in addition to the carboxyl group, are very close. This generates a distribution of negative charges greater than the others. In general, the negative charge density (red lines) for all molecules were lower than the positive charge density (yellow lines), this is due to the FAs structure. These FAs molecules have between 14 and 22 carbon atoms, where the hydrocarbon chain predominates with a negative charge density towards the backbone. Likewise, the negative charge density is only located in the carboxyl group and in the unsaturations.
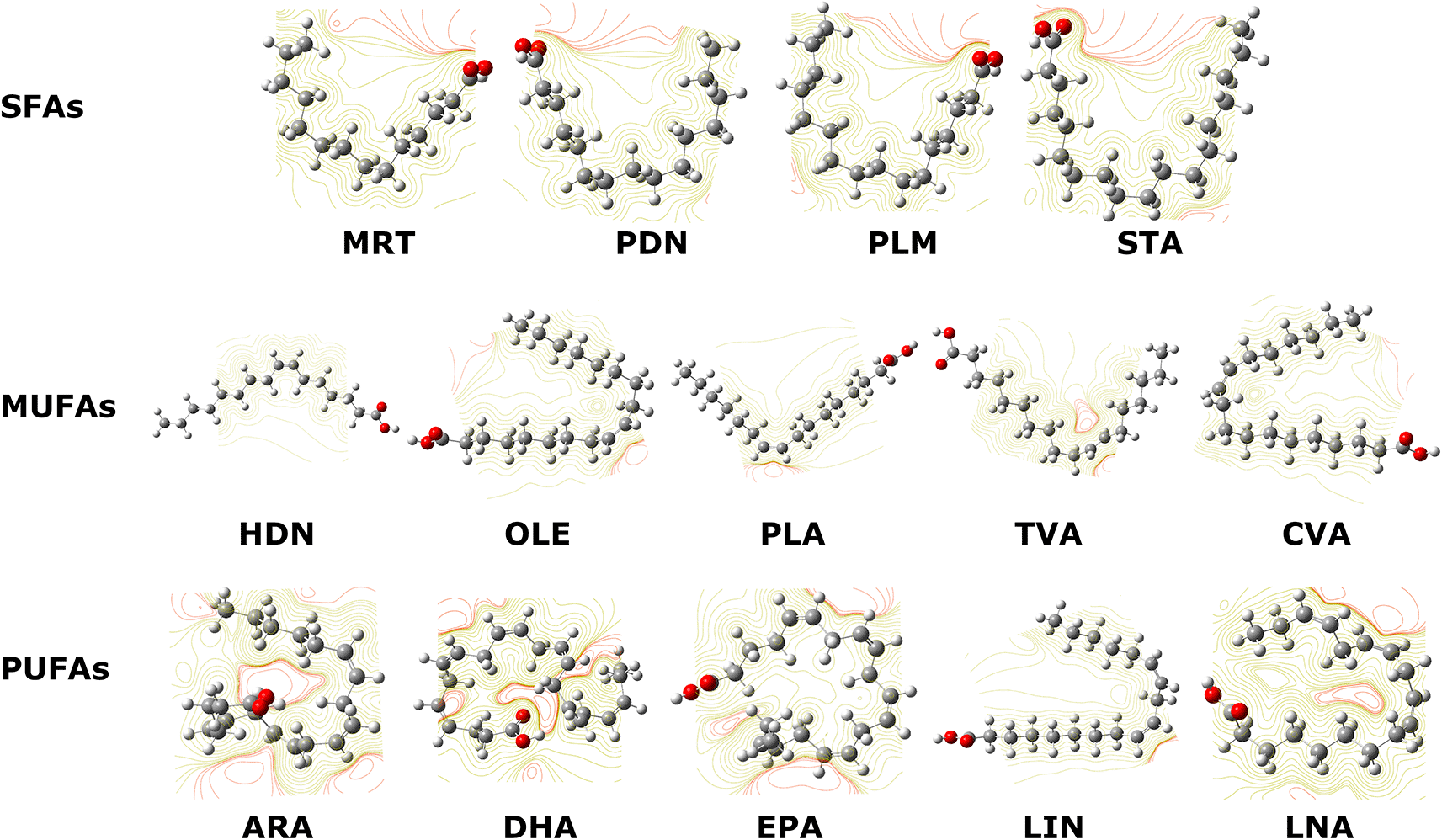
The negative charge density in each atom is represented in red lines and the areas with positive charge density in each molecule are represented in yellow color.
The Fukui function is a local selectivity descriptor.68–70 This parameter reveals the most susceptible molecule area of nucleophilic (), electrophilic (), and radicalary () attacks.68 The Figure 3 shows that DHA, PDN, STA, and TVA molecules were the FAs with areas most susceptible to nucleophilic and electrophilic attack. These compounds have a significant concentration of positive and negative charges; the negative charge density is oriented around the unsaturations (with respect to DHA and TVA) and the carboxyl group.
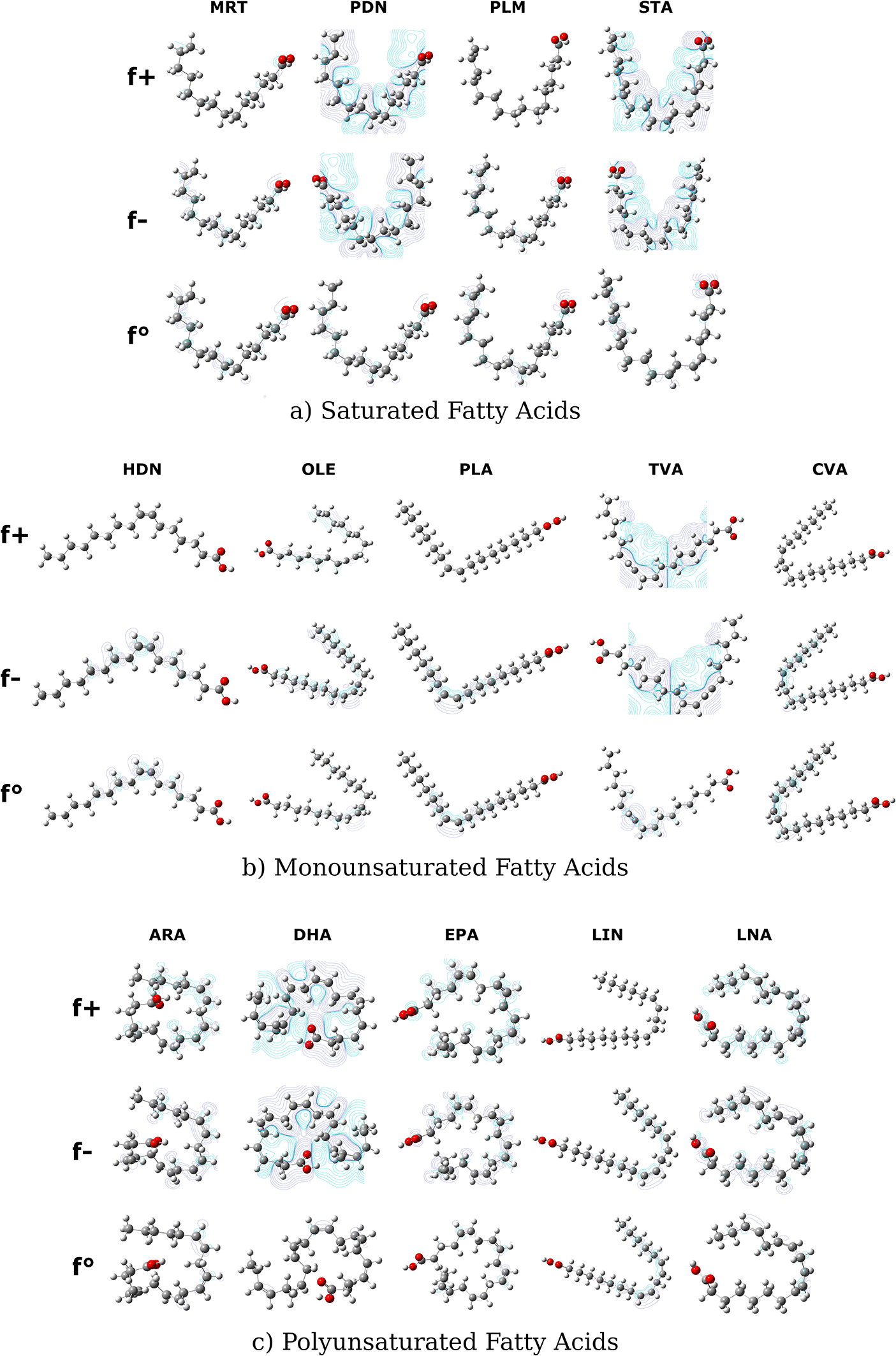
The negative isovalues in each atom are represented in purple lines and the areas with positive isovalues in each molecule are represented in light blue color.
The radicalary attack behavior was different from the and . In this case, the density obtained for all molecules was low. The are focused fundamentally on the unsaturations from FAs. This result is due to fatty acids being susceptible to radical attack by reactive oxygen species in unsaturations.71–75
The molecular weight (WM), number of hydrogen bond donors (nHBD), number of hydrogen bond acceptors (nHBA), and rotatable bonds (nRot) (see Table 4); have shown acceptable values according to the data based on the “Drug-Like Soft” rule (WM(100-600), nHBA(0-12), nHBD(0-7), and nRot(0-11)). The topological polar surface area (TPSA) values of all the compounds reported 37.30 A2, which is an optimal value according to Verber’s rule (140 A2). Therefore, these results show that FAs would have good absorption and permeability. Likewise, all FAs are accepted for Lipinski’s rule of five ((MW500; logP5; hydrogen bond acceptor10; hydrogen bond donors5), indicating acceptable oral absorption and bioavailability.
| Fatty acid | MWa | nHBAb | nHBDc | TPSAd | nRote | logSf | LogD7.4g | Log Po/wh | pKai | Lipinski’s rule | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SFAs | MRT | 228.210 | 2 | 1 | 37.30 | 12 | -4.378 | 3.023 | 5.820 | 4.8 | Yes |
| PDN | 242.220 | 2 | 1 | 37.30 | 13 | -4.814 | 3.135 | 6.283 | 4.8 | Yes | |
| PLM | 256.240 | 2 | 1 | 37.30 | 14 | -5.223 | 3.235 | 6.732 | 4.8 | Yes | |
| STA | 284.270 | 2 | 1 | 37.30 | 16 | -5.879 | 3.449 | 7.571 | 4.8 | Yes | |
| MUFAs | HDN | 254.22 | 2 | 1 | 37.30 | 13 | -4.791 | 3.319 | 6.293 | 4.8 | Yes |
| OLE | 282.260 | 2 | 1 | 37.30 | 15 | -5.559 | 3.549 | 7.131 | 4.8 | Yes | |
| PLA | 254.220 | 2 | 1 | 37.30 | 13 | -4.791 | 3.319 | 6.293 | 4.8 | Yes | |
| TVA | 282.260 | 2 | 1 | 37.30 | 15 | -4.628 | 3.794 | 6.830 | 4.8 | Yes | |
| CVA | 282.260 | 2 | 1 | 37.30 | 15 | -3.308 | 3.447 | 6.169 | 4.8 | Yes | |
| PUFAs | ARA | 304.24 | 2 | 1 | 37.30 | 14 | -5.346 | 4.247 | 6.573 | 4.8 | Yes |
| DHA | 328.24 | 2 | 1 | 37.30 | 14 | -5.384 | 4.803 | 6.699 | 4.6 | Yes | |
| EPA | 302.220 | 2 | 1 | 37.30 | 13 | -5.128 | 4.389 | 5.490 | 4.8 | Yes | |
| LIN | 280.240 | 2 | 1 | 37.30 | 14 | -5.230 | 3.580 | 6.652 | 4.8 | Yes | |
| LNA | 278.220 | 2 | 1 | 37.30 | 13 | -4.973 | 3.690 | 6.156 | 4.8 | Yes | |
The pharmacokinetic properties are reported in Table S1 from Supporting Information. The oral adsorption logP, distribution logD, and solubility logS descriptors were evaluated. The results showed that the logP values are above the desired range (0 to 3 log mol/L), indicating that these compounds are insoluble since they are trapped in the lipophilic bilayer. This could be confirmed by the resulting values of logD (1 to 3 log mol/L) that 197 are between 3 to 5, decreasing the solubility. Furthermore, all the compounds have a pKa = 4.8 except for 7-hexadecenoic acid with a pKa = 4.6. The fatty acids analyzed show optimal values for Caco-2 (>-5.15log cm/s); indicating that they have good permeability to intestinal cell membranes. They are also a substrate for P-glycoprotein (Pgp), where Pgp is a transporter efflux out of cells of xenobiotic molecules.
In general, the FAs have a high percentage (up to 90%) of plasma protein binding (PPB), contrary to DHA and EPA with and , respectively. This union is determined by the structural properties of fatty acids that influence the oral bioavailability of the compounds, where a portion of the free compound could be absorbed through the membranes. The penetration of the blood-brain barrier (BBB) is important for those components that have activity in the central nervous system (CNS), some fatty acids have a better facility to penetrate the BBB than others; the values of lipophilicity, molecular weight, and TPSA allow us to predict a good penetration of the compounds to the BBB.
Cytochrome P450 (CYP) isoforms were also evaluated. The results indicated that fatty acids have a substrate/inhibitory effect on the CYP2C9 isoenzyme, which is important in the oxidation of xenobiotic compounds. ARA, DHA, and EPA fatty acids showed a moderate clearance of 6.011ml/min/kg, 9.988ml/min/kg, and 8.610ml/min/kg, respectively, while the other compounds had values below 5 ml/min/kg, indicating low excretion. Among the results, DHA and EPA present relative toxicity due to their high lipophilicity.
The structure of EgFABP1 was obtained from the Protein Data Bank (PDB), where it has already undergone a rigorous validation process.40 The PDB performs multiple validations, including the generation of a Ramachandran plot, which checks that the (phi) and (psi) torsion angles of the backbone residues fall within allowed or preferred regions.76 This validation ensures that the structure is reliable for subsequent studies. However, for clarity, we have reviewed the structure of EgFABP1 using MolProbity server,77 and confirmed that the torsion angles are consistent with a Ramachandran plot, showing correct conformation with no significant anomalies. The Ramachandran plot analysis of EgFABP1 structure demonstrated that 97% of the residues are in favoured regions, exceeding the typical benchmark for high-quality structures, and 100% of the residues are in allowed regions with no outliers. These results confirm that the protein adopts a highly favourable and physically plausible conformation, indicating that the structure is geometrically valid and reliable for further analysis. This validation reinforces the credibility of the model for subsequent functional and structural studies (see FigureS1 of the supplementary section).
Best protein-ligand binding models were obtained using a genetic algorithm from the Dockthor server. Additionally, an affinity prediction scoring was obtained among the fourteen FAs. This prediction affinity is used to rank different ligands in virtual screening experiments considering the “Total Energy” of each FAs.43,44 According to the score, DHA has a better affinity to EgFABP1 compared to the other FAs (see Table S2 from Supplementary Material).
Regarding Molecular Dynamics analysis, Figure 4 shows the plot of RMSD (root-mean-squared deviation), RMSF (root-mean-squared fluctuation), RG (radii of gyration), SASA (solvent-accessible surface area), and HBond (intramolecular hydrogen bonds from protein) during the simulation time of 200 ns in the isobaric-isothermal ensemble. The analysis of the RMSD plot showed us the MRT, ARA, and EPA with instability from 150 to 200 ns. Similar behaviour has the RMSF analysis, in which low and high fluctuations indicate the structural mobility of each residue in molecular dynamics. The RG plot indicates low compactness between EPA and ARA. While for the SASA study, we observed that the solvent access is similar in coupled systems and apparently, the number of HBonds is reduced in coupled systems.
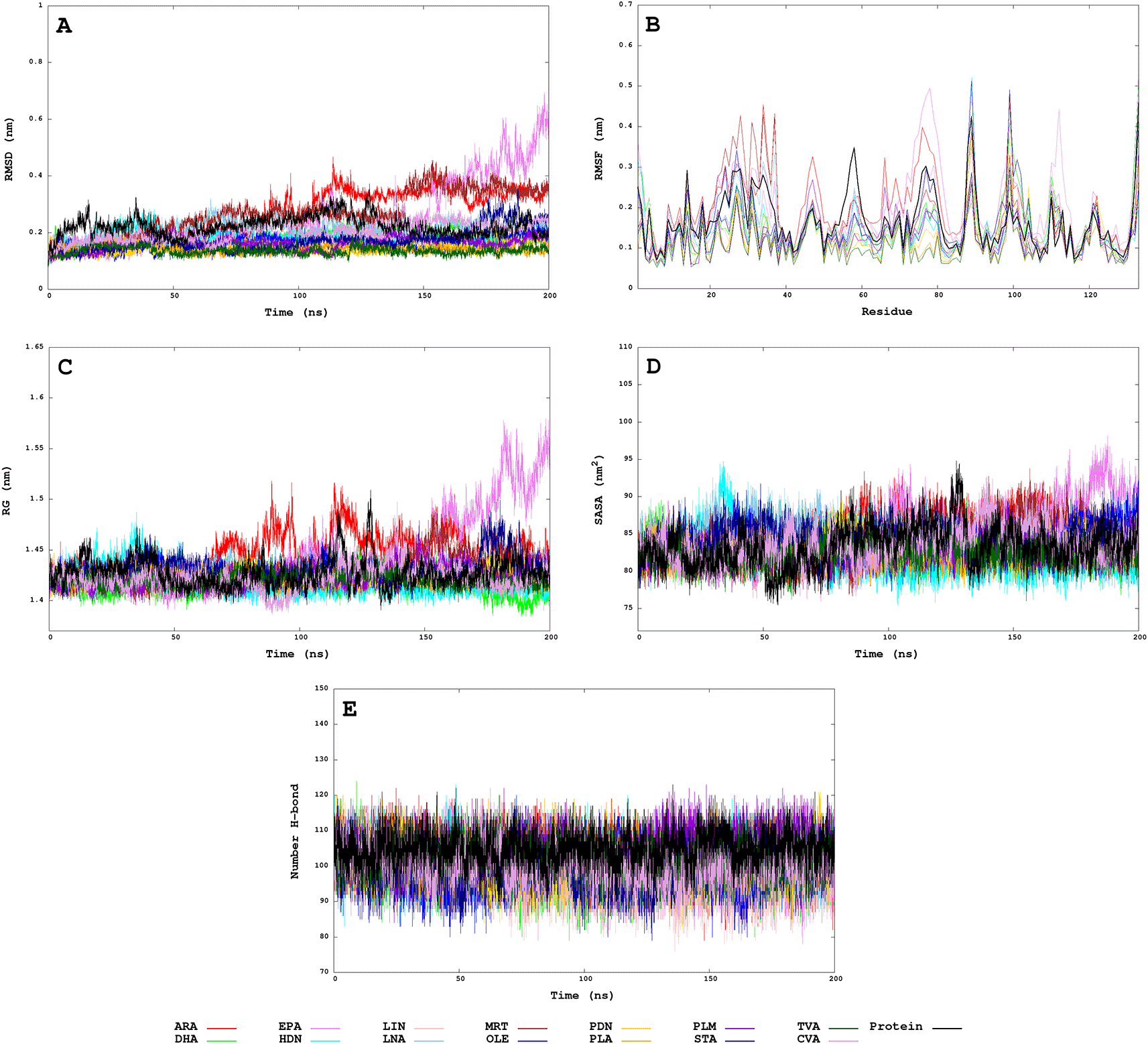
A. Root-mean squared deviation. B. Root-mean squared fluctuation. C. Radii of gyration. D. Solvent-Accessible Surface Area. E. Intramolecular hydrogen bonds from protein.
Table S3 from Supporting Information shows the average values for RMSD, RMSF, RG, SASA, and HBond for the last 20 ns (same time used to estimate binding free energy by MM/GBSA). However hydrophobic interactions and salt bridges were calculated from the last frame. One of the most outstanding systems (receptor-ligand) was EgFABP1-EPA because it presents an average value of RMSD (0.50±0.07 nm), RMSF (0.17±0.08 nm), RG (1.53±0.02 nm), SASA (91.37±2.26) higher and HBond (98.00±5.00) lower than the other systems. The PLA and VAC systems show a lower RMSD (0.14±0.01 nm) and RMSF (0.10±0.04 nm) than the other systems, indicating that the protein is more stable. Additional information about the hydrophobic interactions between EgFABP1-FA are reported in Table S4, Table S5 and Figure S1 from Supporting Information.
The binding free energy estimation was analyzed by MM/GBSA method. Table 5 shows that DHA (-38.52±2.60 kcal/mol G) and PLA (-37.07±4.55 kcal/mol G) obtained the best energy followed by the TVA (-36.52±2.25 kcal/mol G) and OLE (-35.58±3.64 kcal/mol G). All these four ligands are UFAs (i.e. DHA corresponds to PUFAs and the others correspond to MUFAs). These results suggest that the EgFAPB1 prefers unsaturated fatty acids. In parallel, it was observed that the Van der Waals energies give the greatest energy contribution.
On the other hand, EPA obtained lower binding free energy than the other FAs. Figure 5, shows how DHA (better free energy) stabilizes the protein. However, EPA (lower free energy) destabilizes the protein. According to research by Undurti N. Das,78 this effect could be beneficial in overcoming drug resistance against parasitic infections. Other research has shown that DHA has demonstrated direct antiparasitic effects and immunomodulatory properties against several parasitic organisms.13,78 This type of FA can disrupt parasite cell membranes, showing inhibitory effects on Plasmodium falciparum,79 Leishmania,80,81 and Trypanosoma species. DHA also enhances antimalarial drugs such as artemisinin by increasing parasite susceptibility. Its anti-inflammatory effects help regulate host immune responses, reducing excessive inflammation while maintaining effective defence, especially in Trypanosoma cruzi, helminth infections, and Toxoplasma gondii.14 Overall, DHA could be a promising adjunct to antiparasitic therapy for Echinoccoccus granulosus and host immune modulation.
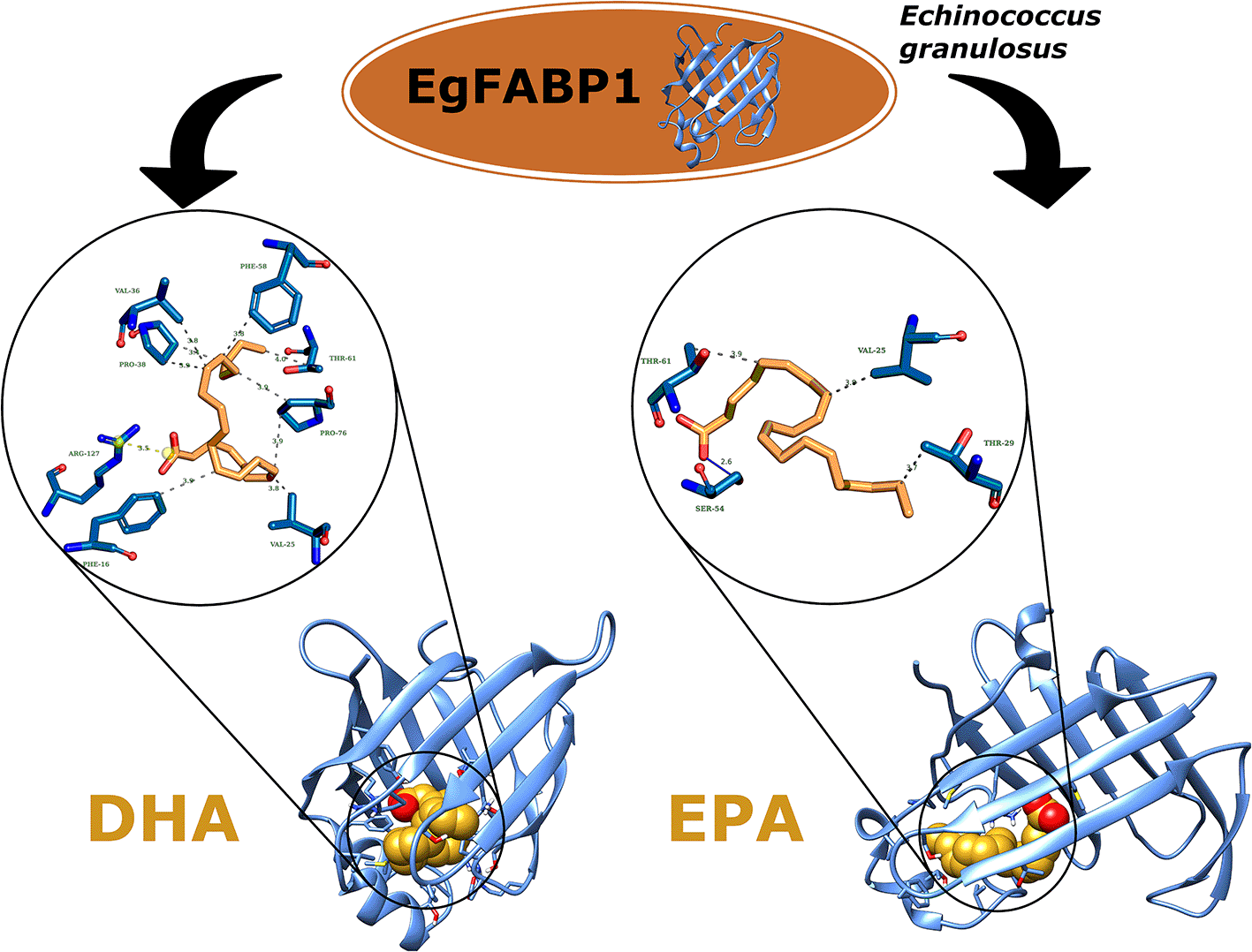
The ligands and active site amino acids are represented in stick form. Hydrophobic interactions between them are indicated by green dashed lines.
Also, we evaluated Spearman’s Rho between RMSD and binding free energy ( TOTAL). In the systems with better free energy, the protein tends to be more stable than without ligand, which could be inversely related. We obtain a Spearman’s Rho = 0.73 and p-value = 0.0029. The results indicate that the relationship between RMSD and binding free energy is a significant (p-value below the usual threshold of 0.05) correlation. In addition, according to most of the research reviewed,82,83 the Spearman’s Rho obtained is classified as a strong correlation.
This study underscores the critical interactions between Echinococcus granulosus fatty-acid-binding protein 1 (EgFABP1) and various fatty acids (FAs), with a particular emphasis on the strong binding affinities exhibited by unsaturated fatty acids such as docosahexaenoic acid (DHA), oleic acid (OLE), and linoleic acid (LIN). The lower binding free energies observed with these unsaturated FAs suggest that EgFABP1 has a marked preference for molecules with double bonds, which may contribute to the parasite’s ability to efficiently manage lipid uptake and metabolism, crucial for its survival and pathogenicity. Saturated fatty acids, including stearic acid (STA) and palmitic acid (PLM), also demonstrated stable interactions with EgFABP1, albeit with higher binding energies, indicating that while EgFABP1 can accommodate a range of FAs, its affinity is notably higher for unsaturated variants. This aligns with research that has shown the structural flexibility provided by double bonds in unsaturated FAs facilitates tighter binding and more stable complexes within lipid-binding proteins. Additionally, the study’s findings with polyunsaturated fatty acids (PUFAs) like eicosapentaenoic acid (EPA) and arachidonic acid (ARA) highlight the potential involvement of EgFABP1 in regulating cellular processes such as membrane fluidity and signal transduction, which are essential for the parasite’s adaptability and virulence. These interactions not only suggest that FAs could serve as valuable metabolic markers but also open the possibility of using FAs as carriers for targeted delivery of therapeutic agents to disrupt the lipid metabolism of E. granulosus. This could pave the way for novel interventions against echinococcosis, particularly in light of the growing interest in exploiting lipid-binding proteins as drug targets in parasitic diseases.
In this study, the chemical reactivity descriptors of fourteen fatty acids were calculated using Density Functional Theory (DFT), and the stability of their complexes was analyzed through molecular dynamics simulations. The objective was to explore the interaction of fatty acids with EgFABP1 from E. granulosus. The lowest HOMO-LUMO energy gap was found in polyunsaturated fatty acids (PUFAs), suggesting greater reactivity. The dipole moment of PUFAs was below 1.5 Debye, implying lower water solubility and an enhanced ability to cross cell membranes. Among these, docosahexaenoic acid (DHA) exhibited the lowest dipole moment (1.0 Debye), indicating high lipophilicity. The molecular dynamics simulation results demonstrated that most fatty acids formed stable complexes throughout the simulation period. However, the EgFABP1-eicosapentaenoic acid (EPA) complex exhibited conformational changes, resulting in an expanded binding pocket and increased solvent exposure. Energy estimation analysis further confirmed that EgFABP1 had favorable binding affinities with the fatty acids, with DHA showing the strongest binding energy at the active site. Overall, this in silico analysis suggests that DHA may have promising potential for the diagnosis and treatment of E. granulosus.
Figshare. Supplementary material, https://doi.org/10.6084/m9.figshare.24939312. 84
This project contains the following underlying data:
• Table 1. Fatty acids abbreviation, PubChem CID and 2D Representation.
• Table 2. Global chemical reactivity descriptors calculated from Koopmans’ theorem.
• Table 3. Values of the global chemical reactivity descriptors calculated from Koopmans’ theorem.
• Table 4. Physicochemical Properties of Fatty Acids.
• Table 5. Calculated MM/GBSA binding free energy of EgFABP1-FAs (kcal/mol).
• Table S1. Chart of values of fatty acids pharmacokinetic properties.
• Table S2. Affinity prediction from docking results.
• Table S3. RMSD, RMSF, RG, SASA and HBond average values, N° Hydrophobic Interactions and Salt Bridges.
• Table S4. Values of hydrophobic interactions between fatty acids and EgFABP1.
• Table S5. Values of salt bridges between FAs and EgFABP1.
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
We appreciate the computational support of the internal project UCSM with grant number 7309-CU-2020. Yoan H. R. Yoan is grateful to the ANID- Postdoctoral 3230141. This work is similar to that existing in a preprint: https://doi.org/10.26434/chemrxiv-2023-z157s.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Tapeworms, antiparasitic analysis, proteomics, recombinant proteins.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: parasitic disease
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Tapeworms, antiparasitic analysis, proteomics, recombinant proteins.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: I work on protein structure and dynamics using NMR spectroscopy and other methods, and I am interested in FABPs.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 21 Nov 24 |
read | ||
|
Version 1 19 Apr 24 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)