Keywords
SPARC tutorial, biological data, 3D mapping, FAIR data, Codeathon
This article is included in the Bioinformatics gateway.
This article is included in the Reproducible Research Data and Software collection.
This article is included in the Hackathons collection.
This article is included in the Python collection.
The Stimulating Peripheral Activity to Relieve Conditions (SPARC) data portal is a platform that allows users to access large sets of curated data and tools. Data on the portal is sourced from researchers working in a range of disciplines applied to peripheral nerve activity, including imaging, computational biology, electrophysiology and clinical research. However, many existing tutorials and examples for SPARC do not document examples of interfaces between tools within the SPARC ecosystem. Thus, we see an opportunity to lower the barrier for entry into the SPARC ecosystem, by creating accessible guides to create workflows using multiple SPARC tools. To do this, we have developed an online hub for hosting tutorials that can guide new users through the entire workflow of acquiring data, processing it, and visualizing it, using common, open-source scientific computing tools and crucially, spanning the repertoire of currently available SPARC tools. Our approach encapsulates the workflow within a Jupyter Notebook, which is designed to be readily accessible to new users of SPARC. Having a hub of tutorials that can be easily understood will not only accelerate the growth of the SPARC community but also increase the uptake and usage of tools by members of the community.
SPARC tutorial, biological data, 3D mapping, FAIR data, Codeathon
This publication is prepared as the outcome of the 2022 SPARC (Stimulating Peripheral Activity to Relieve Conditions) FAIR (Findable, Accessible, Interoperable, and Reusable Codeathon, held 6-8 August. The first edition of this event was held in July 2021. The results of the groups participating in the July 2021 SPARC FAIR Codeathon are collected and accessible on this resource page. In the 2022 SPARC FAIR Codeathon, five groups participated, and more information can be found here.
The goal of the SPARC FAIR Codeathons is to find novel ways of using the SPARC resources, data, or tools to demonstrate and enhance the FAIRness of the data.
The SPARC program is a platform supported by the National Institutes of Health (NIH) Common Fund to advance bioelectronic medicine devices.1 This is done by identifying the spatial distribution of neurons on the surface of organs using flatmaps.2 The SPARC Portal hosts over a hundred datasets and resources (continuously being updated) to assist researchers in learning about organ functions, devise medical tools, and ultimately treat diseases.
The SPARC Portal emphasizes on a FAIR (Findable, Accessible, Interoperable and Reusable) repository of curated resources available to the researchers globally. The FAIR Data Principles were proposed in 2016 by Wilkinson et al. in order to increase the reusability of data.3 The SPARC project incorporates multiple open-source tools and platforms with their own documentation and tutorials. However, well-documented tutorials on how to properly utilize these tools in conjunction with SPARC data are lacking and could significantly aid users in seeing the advantages of using FAIR data.
In this Codeathon we aimed at practically improving the reusability of the SPARC Portal resources by giving visual and written instructions on how the available datasets and models can be employed, integrated, and visualized. One of the core platforms of Data Resource Center (DRC) of the SPARC program is (Open Online Simulations for Stimulating Peripheral Activity to Relieve Conditions).4 Researchers can, for example, use for simulating the peripheral nerve system and instantly observe its impact on pertaining organs. However, the utilization of is not necessarily straightforward for general users. Therefore, to increase the FAIRness of the SPARC Portal resources, we aimed to show how one can search and use the SPARC datasets using available SPARC tools, in conjunction with other open-access tools, in a structured and comprehensible manner. We lower the barrier for the general user who might be unfamiliar with the SPARC integrated platforms (like ) to acquire data, process it, and visualize it using common, open-source scientific computing tools.
The purpose of this article is to present the tutorials that were developed during the 2022 SPARC FAIR Codeathon on the procedure of utilizing the SPARC portal tools and resources. These tutorials demonstrated the ease with which a range of tools in the SPARC ecosystem could be used in concert to access, manipulate and interpret SPARC datasets. As a case study, we provide an example of generating a 3D scaffold of the rat stomach and projecting the coordination data of the vagal afferent and efferent. We discuss how this tutorial differs from the existing tutorials and documentation on SPARC Portal. Finally, we describe how our tutorial style can impact the FAIRness of the SPARC Portal and possible future developments.
This section discusses the improvement of tutorials and documentation on the utilization of the SPARC Portal tools along with the available open-access software packages such as SciPy. Figure 1 demonstrates the general workflow for the tutorials. The first step is to obtain SPARC datasets from the portal using Pennsieve. This free tool is a scalable, graph-based, and cloud-based data management platform that is used for managing scientific datasets on the SPARC Portal. It is used here to download.xlsx files. Once the files are downloaded from a project, the data can be manipulated as needed in order to perform computational analysis on it by employing a programming language such as Python or open-source tools from the SPARC Portal. After the processing step, the data can be visualised in 2D or 3D graphs. In the following, we explain these steps and the cases demonstrated in the tutorial.
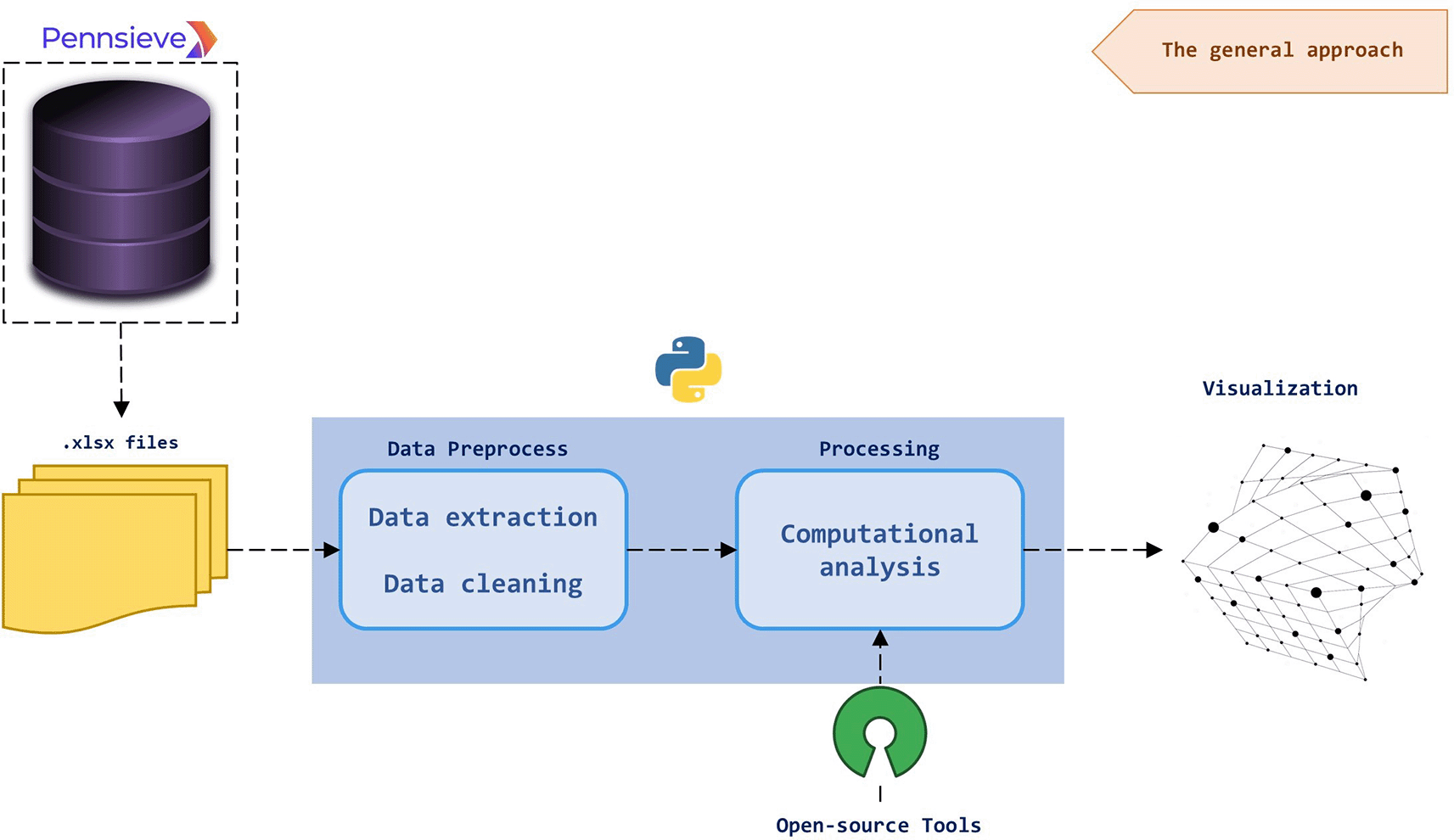
The.xlsx data files are downloaded using Pennsieve and fed to a program written in Python which uses open-source tools, like those available from SPARC, for preprocessing and analysing the data. After being processed, the data can be visualised in different manners.
Figure 2 illustrates the above-mentioned workflow using a previously published model of the rat stomach.5 We illustrate two applications of the acquired datasets and give detailed instructions on how they can be processed for different applications. Three datasets constitute the basis of these tutorials. They provide the 2D coordinates for different neuronal populations in the rat stomach, namely the efferent vagal neurons and afferent vagal neuron sub-populations of intraganglionic laminar endings (IGLEs) and intramuscular arrays (IMAs). These data are derived from biomedical imaging of stained rat stomach sections. The information regarding these neurons is contained in.xlsx files that are accessible on the SPARC portal from the following links (see also Underlying data):
• Spatial distribution and morphometric characterization of vagal afferents associated with the myenteric plexus of the rat stomach: IGLE_data.xlsx
• Spatial distribution and morphometric characterization of vagal efferents associated with the myenteric plexus of the rat stomach: Efferent_data.xlsx
• Spatial distribution and morphometric characterization of vagal afferents (intramuscular arrays (IMAs)) within the longitudinal and circular muscle layers of the rat stomach: IMA_analyzed_data.xlsx
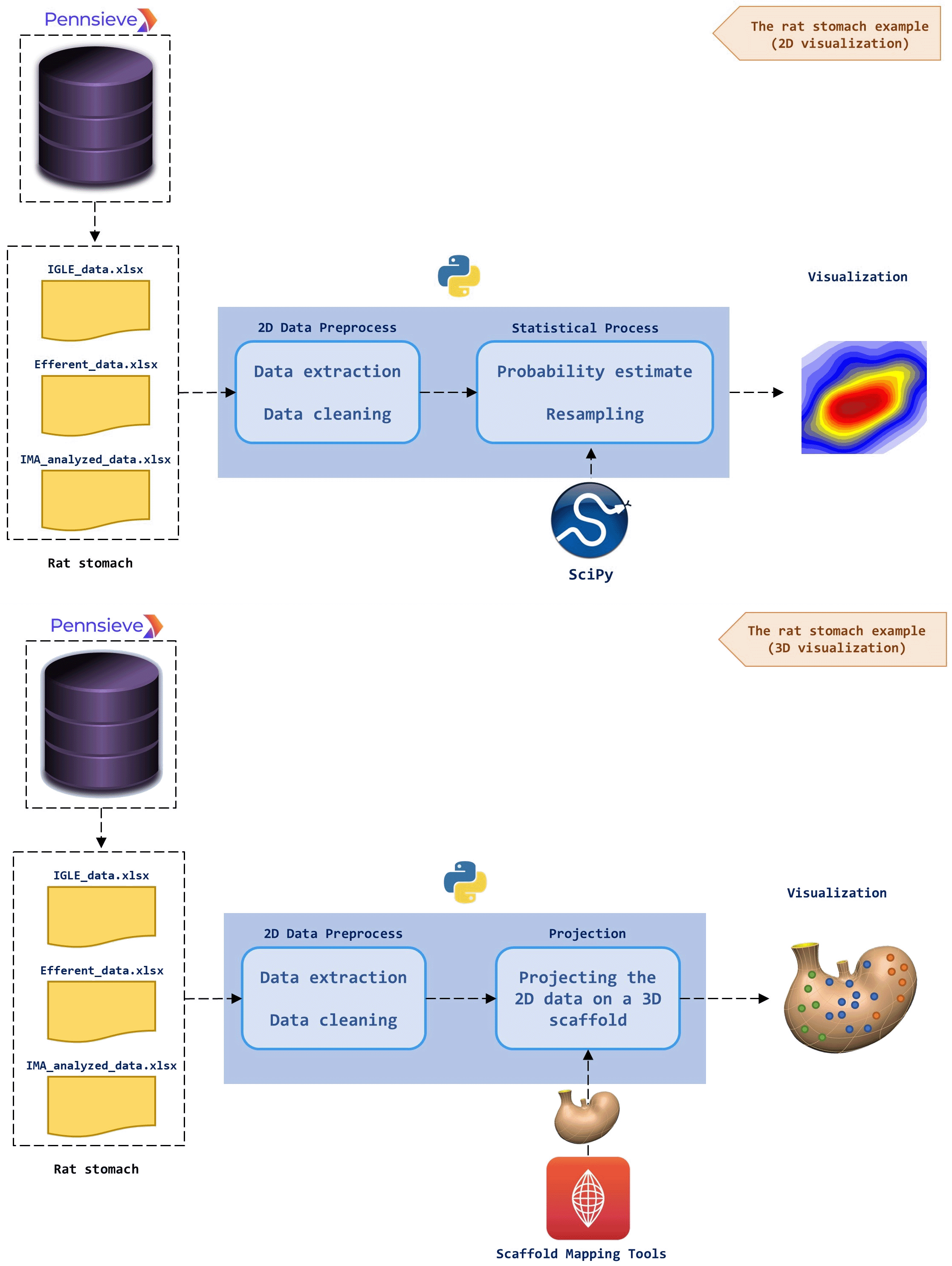
The upper panel (the general approach): The.xlsx data files are downloaded from Pennsieve and fed to Python for preprocessing (data extraction and cleaning). Using the available Scaffold Mapping Tools on the SPARC Portal, a geometric 3D scaffold of an organ is obtained. The preprocessed data can be projected on the 3D scaffold to create a data-enriched 3D image of the organ. The lower panel (the rat stomach example): Three different datasets of the spatial distribution of neurons in the rat stomach are downloaded from Pennsieve. The required 2D data (spatial coordinates of x and y) are extracted using Python. A 3D scaffold of the rat stomach is achieved using the SPARC Scaffold Mapping Tools. The preprocessed data of the spatial 2D distribution of neurons is projected on the rat stomach scaffold. The output is a 3D image of the rat stomach in which the neurons are mapped on the surface. The dots correspond to the spatial distribution of neurons and the three colors represent the three different datasets.
In this tutorial, we demonstrate a dataset acquisition mechanism that includes searching for datasets using keywords, e.g., ‘vagal,’ using the search API provided by Pennsieve with output datasets’ id, version, name, and tags. Using the search results, we can now download datasets manually or automatically, where the automatic method is facilitated by a script utilizing the discovery API that can be accessed freely. This mechanism is then helpful in searching for other datasets, which will later be used for different analysis purposes.
The datasets obtained can be in various formats, for example, JSON, dat, knowledge-based, and xlsx; for the cases we raised, the file is xlsx. Within these datasets, we conserve information regarding the neuron location, innervation area, and anatomical location, whether dorsal or ventral. The neuron’s location is a 2D feature representing the percentage relative distance to an origin situated in the pyloric end of the stomach for the y-axis (left to right direction) and near the esophageal for the z-axis (bottom to top direction). To be able to plot this location, first, we define the boundary of z-axis and y-axis based on the 3D stomach scaffold generated using the Scaffold Mapping Tools of SPARC, and again in our cases we set to [0, 36.7] and [0, 24.6] respectively, where each element is the minimum or maximum point in millimeter. This boundary can calculate the approximate neuron location in the millimeter.
After data manipulation, one can use location data to estimate the probability density of the spatial distribution of neurons in 2D as shown in the upper panel in Figure 2. For this visualization, first, we create a 2D mesh grid with z-axis and y-axis boundaries consisting of 100 points each. Then for each neuron location, the probability density is estimated using Gaussian KDE (Kernel Density Estimation)6,7 and resampled by 1000 points. We estimate all three datasets and visualize them in 2D plots. It is interesting that estimates can use methods other than KDE for different analysis needs and datasets, such as the Cauchy and Laplacian kernels.
Moreover, one can project the 2D points representing the location of neurons to a scaffold of the organ generated by the Scaffold Mapping Tools of SPARC as demonstrated in the lower panel of Figure 2. The scaffold organ obtained is a 3D mesh converted as an STL file to handle it using a Python-based package easily. Figure 3 is the representation of the 2D dataset that we map to the 3D organ scaffold. 3D mapping can be done because the datasets contain information on the anatomical location of neurons, dorsal and ventral.
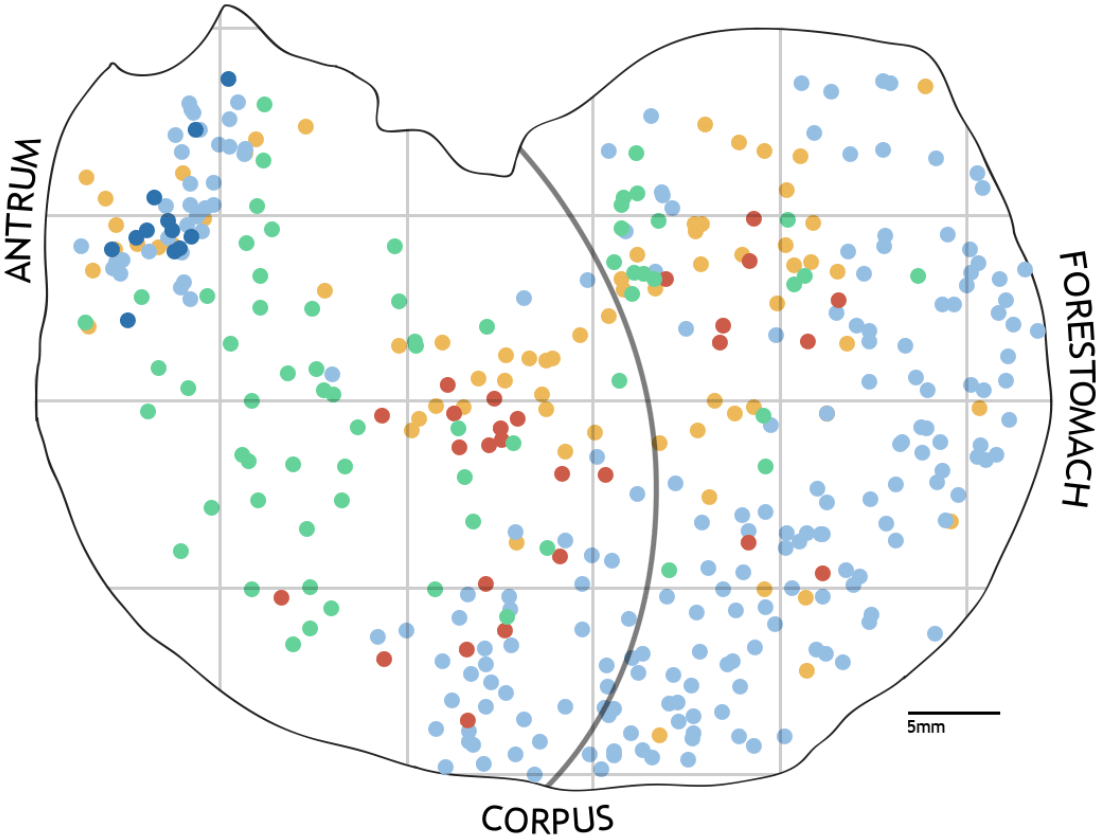
The color difference indicates the type of neuron, and the color intensity indicates the area of the neuron (retrieved from [1]).
The step-by-step instructions on how to use the Python code are available on GitHub (see Software availability).
In this section, we demonstrate the graphical results from the two tutorials that were developed during the Codeathon.
Figure 4 illustrates the results of the first tutorial, data resampling for density estimation simulations using KDE. The simulation includes all three datasets: vagal efferents, IGLEs, and IMAs. Within the plots, the green dot signifies the resampled point regarding vagal, while the backdrop color denotes the density of the data points. Areas in dark red indicate high density, while lighter ones represent lower density. It can be seen here that the highest concentration of vagal efferents is at 50% and above the x-axis approaching the forestomach. As for IGLEs, the distribution is relatively even, with the highest density between 10% and 50% of the x-axis. Meanwhile, IMA is concentrated in the antrum and scattered in the proximal stomach. These plots confirm the distribution of the stomach’s vagal afferent and efferent populations.5 They could be an alternative to visualize the distribution more effectively and efficiently. Here, we demonstrate that the datasets and tools provided by SPARC can be easily used and combined with other open-source tools. This tutorial module does not consider dorsal and ventral positions, naturally allowing for more precise visualization. We provide this space for learners as a learning process.
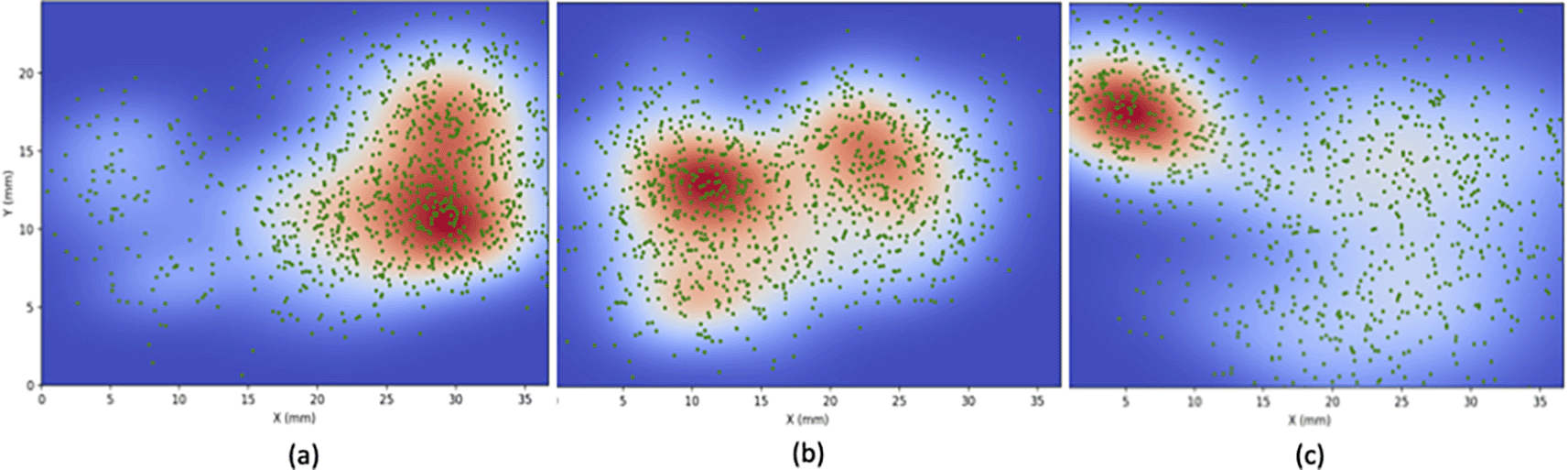
(a) efferent (b) IGLE (c) IMA. Statistical processes are used to compute a probability density estimation of neurons. This density function can be used to generate and simulate neuronal activity in a network.
Figure 5 illustrates the 3D mapping of the spatial distribution data to the rat stomach scaffold. The datasets for this module are the same as for the previous module, using data features on the ventral and dorsal position as well as the area of innervation. With this visualization, we can see the extent of innervation and later compare it with a 2D density plot. This area is tiny; the magnitude plot is inefficient; therefore, it can be represented well using a colour scale from white to dark green, where dark green represents the most significant area. We used the matplotlib widget module for8 interactivity so that the 3D plot could be rotated to observe the distribution of the vagal and area innervation in more detail.
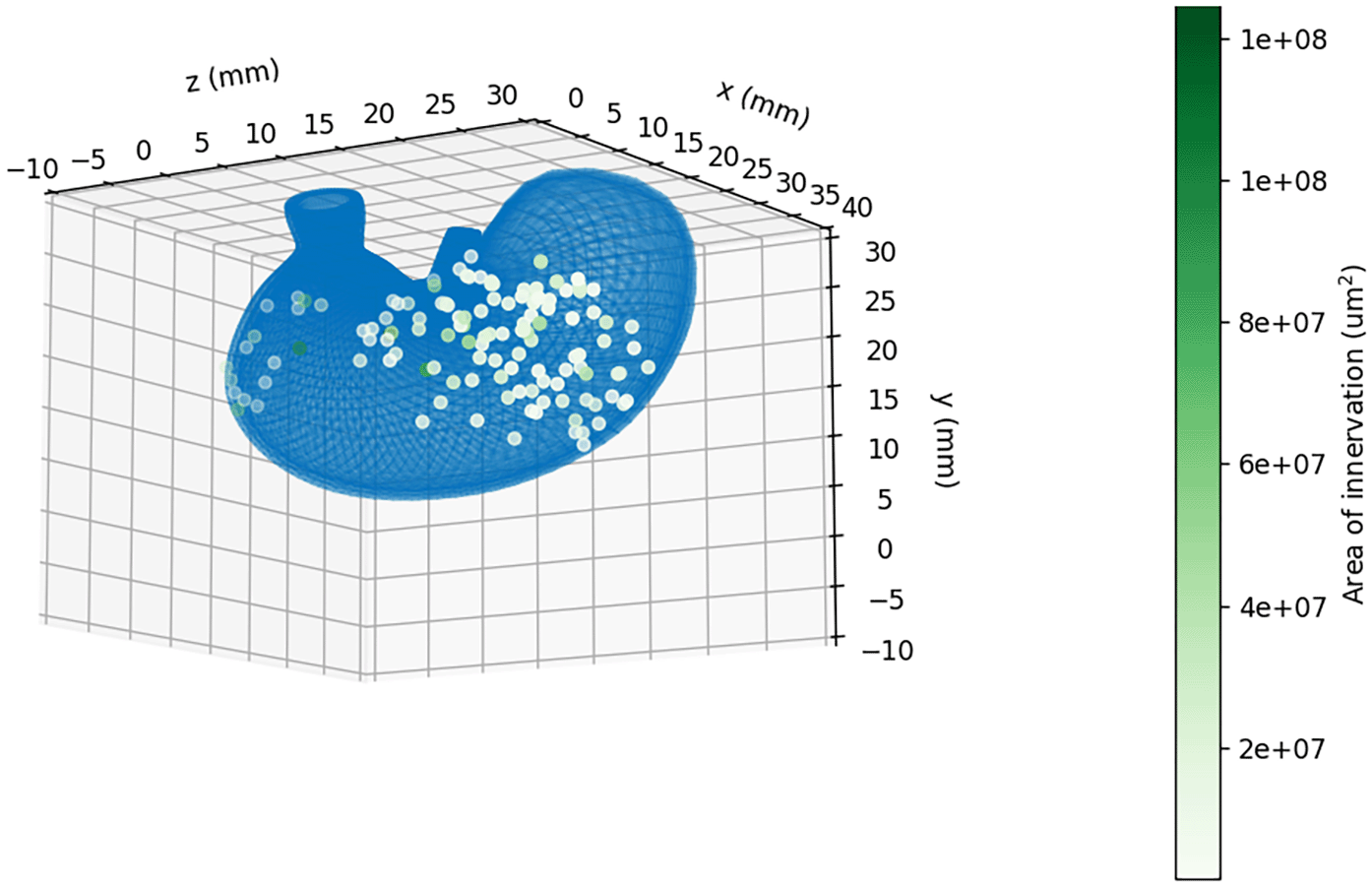
The result of our tutorial is the visualization of the neurons in an interactive figure in Jupyter Notebook. The user can select which type of neurons to visualise on the surface of the stomach from three categories: vagal afferent (IGLE), vagal afferent (IMA), and vagal efferent.
Our tutorials differ from the current tutorials on the SPARC Portal in a substantial way because they guide the users through understanding and using multiple SPARC tools to fulfil data manipulation and visualisation needs. Instead of the dense documentation and tutorials on the SPARC portal which mostly targets a single tool and is appropriate for users already familiar with the SPARC program, we have developed tutorials, along with examples, for a broader audience. We demonstrated the feasibility of using SPARC resources to drive our proposed workflow using a rat stomach scaffold as an example. A central benefit of this approach is that it leverages the FAIR principles that underlie these SPARC resources to ensure that the integrated models that are produced also follow the FAIR Data Principles. Our approach is creative because it encapsulates the workflow within a Jupyter Notebook online platform, which is designed to be readily accessible to new users of SPARC. Having tutorials that can be easily understood will not only accelerate the growth of the SPARC community but also increase the uptake and usage of tools by members of the community.
The data for the rat stomach example presented as a use case for our tutorials is available from SPARC at the following links:
https://sparc.science/datasets/10?type=dataset&datasetDetailsTab=abstract&path=files%2Fderivative
https://sparc.science/datasets/12?type=dataset&datasetDetailsTab=files&path=files%2Fderivative
https://sparc.science/datasets/11?type=dataset&datasetDetailsTab=files&path=files%2Fderivative
Source code available from: https://github.com/SPARC-FAIR-Codeathon/QuiltedTutorials
Archived source code at the time of publication: https://doi.org/10.5281/zenodo.8323579. 9
License: MIT
Visit our website to find an expanding collection of tutorials: https://quilted-tutorial.github.io/SPARC-guru
The archived source code at the time of publication: https://doi.org/10.5281/zenodo.8323579. 9
All authors were part of the team QuiltedTutorials in the 2022 SPARC FAIR Codeathon. We would like to extend our special thanks to the SPARC Program of the NIH Common Fund and to the organisers of the 2022 SPARC FAIR Codeathon for their support during the planning and development of this project.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new method (or application) clearly explained?
Yes
Is the description of the method technically sound?
Yes
Are sufficient details provided to allow replication of the method development and its use by others?
Yes
If any results are presented, are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions about the method and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: calcium imaging in different models and cell types, data analysis, reproducible data analysis and training in python-based data processing
Is the rationale for developing the new method (or application) clearly explained?
Yes
Is the description of the method technically sound?
Partly
Are sufficient details provided to allow replication of the method development and its use by others?
Partly
If any results are presented, are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions about the method and its performance adequately supported by the findings presented in the article?
Partly
References
1. Bandrowski A, Grethe J, Pilko A, Gillespie T, et al.: SPARC Data Structure: Rationale and Design of a FAIR Standard for Biomedical Research Data. bioRxiv. 2021. Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Data standards, FAIR practices, Research software
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 08 Jan 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)