Keywords
Pseudomonads, multidrug resistance, environmental reservoirs, carbapenem-resistant Pseudomonas aeruginosa
This article is included in the Antimicrobial Resistance collection.
Pseudomonads are gram negative bacteria and readily form biofilms in the environment, allowing long-term colonization and persistence in sinks, water systems. They pose a risk of life-threatening opportunistic infections in immune-compromised individuals. MDR strains, make treatment increasingly difficult. Environmentally persistent MDR strains are typically problematic within healthcare facilities, however, data on MDR pseudomonad reservoirs in settings with community-acquired infections to inform preventive interventions, in resource-constrained settings is scarce. Here, we determined reservoirs and antibiotic susceptibility of Pseudomonas species in water sources in Kisumu County, Kenya with reported high levels of community acquired pseudomonad infections.
We adopted a cross-sectional design, randomly collecting 297 samples from tap heads, sinks, tanks, vendor and household storage containers in six selected sub-locations and one hospital (KCRH). Standard microbiological procedures were used for identification and AST of the isolates.
We isolated Pseudomonads from 14.1% of the samples collected, predominantly from the community 10.4%. Seven different pseudomonads were identified, with Pseudomonas aeruginosa predominating 6.7% overall, in the community samples 5.7%, and among isolates from water tanks 21.4%. Pseudomonad isolates were 62% non-susceptible to piperacillin, 57% to tigecycline, 24% meropenem, 21% cefepime, 19% levofloxacin and 14% colistin. Carbapenem resistance was mainly detected in P. aeruginosa 80% (8/10) from Milimani sub-location 75% (6/8). 45% of the isolates recovered were MDR, mainly community-associated carbapenem-resistant P. aeruginosa (CRPA) 42%, strains susceptible to colistin. The MDR pseudomonads exhibited high multiple antibiotic resistance indices, ranging from 0.43 to 1.
This study reveals a higher prevalence of MDR pseudomonads, including CRPA strains in community water sources. These potential conduits of drug resistance present a critical public health threat, especially among immunocompromised. Regular cleaning of water storage facilities, water treatment and implementation of antimicrobial stewardship programs, are required to prevent a rise in AMR and eliminate the environmental reservoirs that put the vulnerable populations at risk.
Pseudomonads, multidrug resistance, environmental reservoirs, carbapenem-resistant Pseudomonas aeruginosa
Pseudomonads are gram-negative bacteria, ubiquitous in water soil, and decaying vegetation and frequently isolated from healthy persons’ skin, throat and stool. They comprise several clinically relevant species, including P. maltophilia, P. fluorescens, P. cepacia, P. stutzeri, P. putrefaciens, and P. putida,1,2 with P. aeruginosa as the most common among these opportunistic pathogens. Pseudomonads form biofilms that confer protection from environmental stresses, including antibiotics and detergents, allowing long-term colonization and persistence in sinks, municipal drinking water systems, hot tubs, and swimming pools.3,4
Pseudomonads pose a substantial risk of acute and chronic life-threatening multidrug-resistant (MDR) infections such as ventilator-associated pneumonia, community-acquired pneumonia, bloodstream infection, urinary tract infections, and skin and soft tissue infections among elderly individuals and those with cystic fibrosis, cancer, HIV, and diabetes.5–9 Outbreaks of P. aeruginosa infections have been associated with tap water,10 bottled water,11 hand-washing basin water,12 surface cleaning equipment,13 therapy equipment such as catheters, humidifiers,14 sinks,15 and swimming pools.8
Treatment of infections caused by Pseudomonas species is increasingly difficult due to the rapid emergence of multidrug resistance, even during therapy courses.16 The resistance occurs mainly through reduced outer membrane permeability, expression of efflux pumps that transport antibiotics extracellularly, biofilm formation, and the production of beta-lactamases, and occasionally plasmid-borne enzymes. Of these enzymes, carbapenemases pose a public health concern because they inactivate carbapenems, which are among the antibiotics of last resort for infections caused MDR Gram-negative bacteria (GNB).17,18
MDR P. aeruginosa clinical strains that are carbapenem-resistant (CR) are increasingly reported in Kenya19–21 The available treatment options for CR-GNB include colistin and tigecycline, with colistin rarely used due to high toxicity and low efficacy concerns. Increased resistance to both drugs is growing with the emergence of resistant strains. Newer options for infections caused by CR-GNB include combinations of beta-lactam-beta-lactamase inhibitors (BL-BLI) such as meropenem-vaborbactam, ceftazidime-avibactam, imipenem-sulbactam, and ceftolozane-tazobactam22; however, these drugs face challenges due to insufficient high-quality clinical data, delayed approval for susceptibility testing methods, antibacterial spectra complexity, and acquisition costs.23 Therefore, preserving the clinical value of carbapenems through local, global and national mitigation strategies remains critical.
Although Pseudomonas spp. are significant clinical pathogens,21,24–26 information on the occurrence of MDR isolates in water sources and moist environments in community and hospital settings which could serve as reservoirs for human infections in Kenya is scarce.27 In this study, we aimed to identify reservoirs and antibiotic susceptibility of Pseudomonas spp in community and hospital environments in Kisumu County where community-associated infections have been reported at the county referral hospital.
Approval to conduct this study was obtained from the Kenyatta University’s Graduate School, Kenya Medical Research Institute-Scientific Ethics Review Unit (KEMRI/SERU/CCR/0190/4117 dated February 15th 2021) and the National Commission for Science, Technology and Innovation (NACOSTI/P/20/4221). Verbal consent was obtained and it was approved by KEMRI/SERU to collect samples from households, water distribution points owners and Kisumu County Referral Hospital management.
We conducted this study in the Kisumu County Referral Hospital (KCRH) and six sub-locations within Kisumu County (Figure 1), with the sites chosen based on data from an ongoing study by Musila et al., on antimicrobial resistance in military and civilian populations in Kenya (SSC 2767 WRAIR 2089), where it was observed that the majority of Pseudomonas aeruginosa infections were community-acquired (unpublished data).
We adopted a cross-sectional study design with a random sampling strategy to obtain 297 (100 water and 197 swab) samples from KCRH and six sub-locations; Milimani, Nyalenda A, Nyalenda B, Manyatta, Manyatta B, and Kaloleni within two (2) weeks in April 2021. Seven households, two public schools and two communal water distribution points from each sub-location were randomly selected and 77 water and 172 swab samples from taps, shower heads, tanks (>1000 litres external), water storage containers (20 liters) and sinks were collected. In KCRH, a total of 48 samples were collected from taps, sinks and shower heads in the male general, surgical, pediatric and maternity wards, and from water reservoir tanks.
We collected water samples in clean and sterile screw-capped 250 mL GosselinTM polypropylene bottles containing 0.1 N of 30mg sodium thiosulphate (Thermofisher scientific Catalogue No: 15103917) by disinfecting the tap using 70% ethanol (plastic taps) or flaming (metal taps), and allowing the water to run for a few minutes before sampling. Swab samples of tap outlets, sink basins, shower heads, and interior of tanks and containers were collected using an environmental sampling kit (Puritan ESK, Guilford, ME, USA) as described by Schiavano and others.8 All samples were transported in a cool box (4-6°C) to the KCRH -based USAMRD-A laboratory in Kisumu for processing within 2 hours.
We processed the study samples following standard procedures outlined by the UNI EN ISO 16266:2008 standard on the detection and enumeration of P. aeruginosa method by membrane filtration.28 Briefly, 100 mL of water was filtered through 0.45 μm, 47mm diameter cellulose ester membrane (MF-Millipore, Merck, KGaA, Darmstadt, Germany) to concentrate the bacteria, and the membranes were placed directly onto CHROMagarTM Pseudomonas media (Becton Dickinson, France) and aerobically incubated at 37°C for 18 hrs. Blue or green pyocyanin-producing colonies were considered presumptive P. aeruginosa and fluorescent non-pyocyanin-producing colonies as other pseudomonads. The colonies were sub-cultured on Mueller Hinton Agar (MHA) Nutriselect® Plus (Merck KGaA, Darmstadt, Germany) and screened by Gram staining and oxidase testing. All the oxidase-positive isolates were identified on the VITEK 2® automated platform (bioMèrieux, Marcy I’Etolie, France) using the GN-ID card, and the O-Acetylase antigen-based PCR29 was used to distinguish P. aeruginosa from other pseudomonads.
The VITEK 2 AST-XN05 card was used for antimicrobial susceptibility testing (AST) and interpreted based on the clinical laboratory standards institute guidelines of 2022.30 P. aeruginosa ATCC 27853 and E. coli ATCC 25922 were used as positive and negative controls, respectively. The antibiotics panel included ticarcillin/clavulanic acid (8/2-128/2 μg/ml), piperacillin (4-128 μg/ml), ceftriaxone (0.25-64 μg/ml), cefepime (0.12-32 μg/ml), meropenem (0.25-16 μg/ml), levofloxacin (0.12-8 μg/ml), tigecycline (0.5-8 μg/ml) and chloramphenicol (4-64 μg/ml) on (AST-XN05, 26247, bioMèrieux, Marcy IÉtolie, France). For the colistin AST, the standard Cation adjusted Muller Hinton broth (CAMHB) method was used as described in the CLSI guidelines, with a known mcr-1 positive E. coli and ATCC 27853 as positive and negative control organisms, respectively.
We captured, compiled and analyzed the study data using MS Excel spreadsheet (Microsoft office 2019)31 bar graphs were generated on the same software. This study defined MDR pseudomonads as isolates resistant to 3 or more classes of antibiotics,28 and we calculated the multiple antibiotic resistance indexes (MARI) as described by Davis and Brown32: a/b where ‘a’ was the number of antibiotics to which an isolate exhibited resistance against those it was exposed to ‘b’, for susceptibility.
The highest proportion of samples were collected from the community (83.8%, n=249), drawn mainly from water storage containers (33%, n=98) and taps (24%, n=72) distributed across the different sub- locations and KCRH hospital in Kisumu county (Table 1).
In this study, we isolated pseudomonads from 14.1% (42/297) of the samples collected, predominantly from the community samples (10.4%, 31/297), and identified seven7 different species namely; P. aeruginosa, P. fluorescens, P. putida, P. fluorescens, P. oleovarans, P. mendocina and P. stutzeri. Pseudomonas aeruginosa was the most common species overall (6.7%, 20/297) and in the community samples (5.7%, 17/297). P. mendocina, P. alkaligens and P. putida were only detected in the community samples. P. fluorescens (2.0%, 6/297) was the main species in the hospital samples and P. oleovarans was exclusive to the hospital (Figure 2).
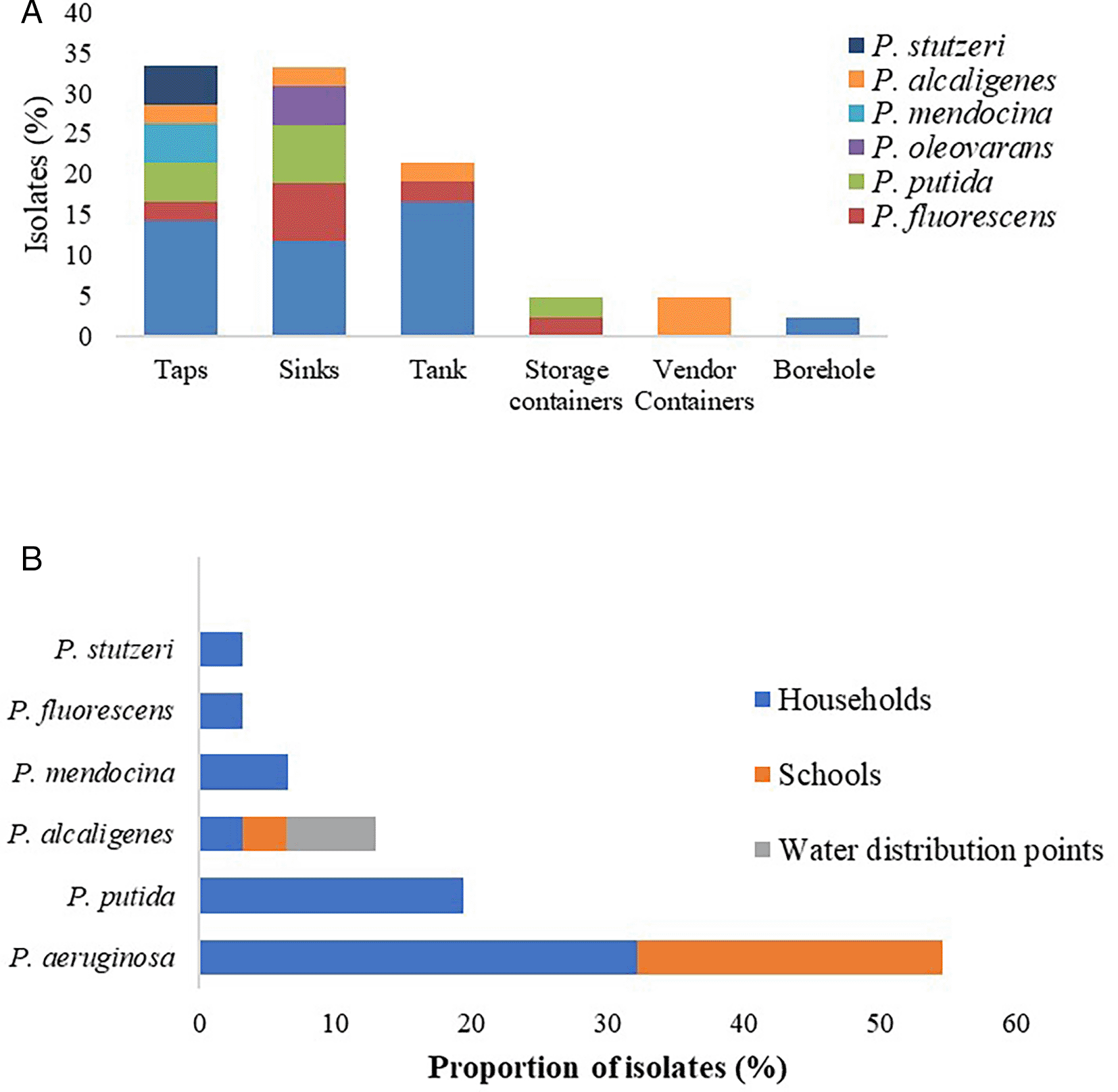
We found pseudomonads in all the study sampling sources, predominantly in taps and sinks (33.3%, 14/42). P. aeruginosa was the most common species in tanks (21.4%, 7/42) but was not isolated from vendor containers, household storage containers, or water distribution outlets, Figure 3A.
Generally, the prevalence and distribution of Pseudomonas spp. varied in all the study sub-locations. The highest prevalence was observed in Milimani (35.7%, 15/42), KCRH (26.2%, 11/42), and Manyatta B (19%, 8/42). P. aeruginosa was predominant in Milimani (28.6%, 12/42), with a few isolates from KCRH and Manyatta but was not isolated in Nyalenda A, Nyalenda B, and Manyatta A (Figure 3B). The greatest diversity of Pseudomonas spp. was observed in taps, KCRH and in Kaloleni sub-location.
Households accounted for 67.7% of the 31 pseudomonads isolated in the community environment, with the majority from taps (32.3%), sinks (22.6%), and tanks (19.4%). P. aeruginosa was the predominant species, recovered mostly from sinks (16.1%) and taps (12.9%) but absent in vendor containers, boreholes, and tank samples. Similarly, P. aeruginosa was the leading isolate (16.1%) in schools, within taps and boreholes (Figure 4).
In the hospital environment, the majority of pseudomonads (36.4%, 4/11) were isolated from swab samples from the tank (27.3%, 3/11) and tap water (9.1%, 1/11). Tank swabs were the only source of P. aeruginosa in hospital samples, Figure 5.
Pseudomonads isolates were 62% (26/42) non-susceptible to piperacillin, 57% (24/42) to tigecycline, 21% (9/42) to cefepime, 19% (8/42) to levofloxacin and 14% (6/42) to colistin (Table 2). Carbapenem resistance (non-susceptibility to meropenem) was 24% (10/42), mainly recorded in P. aeruginosa (80%, 8/10) from the Milimani sub-location (75%, 6/8). Six isolates (6/42 15%), were non-susceptibility to colistin and were mainly P. putida (50%, 3/6), with P. fluorescens (17%, 1/6) exhibiting resistance to both meropenem and colistin. A single isolate grew poorly during the VITEK 2 AST analysis and were indicated as a ‘terminated result’ (TRM) with 3 repeats.
In this study, 45% (19/42) of the isolates were MDR, predominated by carbapenem-resistant P. aeruginosa (CRPA) (42%, 8/19) and carbapenem-susceptible P. putida (26%, 5/19) mainly from the community samples (Table 3). All MDR CRPA remained susceptible to colistin and were isolated from the Milimani sub-location (80%, 8/10). P. oleovarans, P. alcaligens, P. mendocina, and P. stutzeri were non-MDR, whereas MDR P. putida were carbapenem-susceptible. MDR-pseudomonads exhibited high multiple antibiotic resistance indices (MARI), ranging from 0.43 to 1.
Pseudomonas aeruginosa is a well-studied opportunistic Pseudomonad with a predilection to cause infections in vulnerable populations such as immunocompromised individuals. The global rise in MDR P. aeruginosa, particularly the carbapenem-resistant strains, pose a critical public health challenge due to limited treatment options. In our study setting in Kisumu county, Kenya, P. aeruginosa and other pseudomonads were previously isolated from patients who acquired them in the hospital and in the communities, prompting the need to identify their environmental reservoirs and antibiotic susceptibility patterns to inform the design of targeted infection prevention and control (IPC) interventions both in the community and hospital environments. This study identified pseudomonads in the water sources from the community (schools, households, water distribution points) and the KCRH hospital within Kisumu county at a rate of 14% (42/297). In these water sources P. aeruginosa was the most prevalent species (6.7%), being isolated from tanks (19%, 7/36), borehole (17%, 1/6), sinks (16%, 6/37) and taps (6%, 6/98). A study by Opperman33 reported Pseudomonas in stagnant water, which presents a conducive environment for its proliferation through formation and dissemination of biofilms.34 In the current study, we did not recover P. aeruginosa from water storage containers both in homes and from water vendors, probably due to the shorter static period of water in these containers, considering that these containers were used for ferrying water as opposed to holding it for long periods. Colonization at the point of use, including tap heads, plays a critical role in P. aeruginosa infections as opposed to the water distribution system.35 These data suggest that community interventions should be focused on long- term storage receptacles and piping systems that terminate in tap heads.
In the community setting, the occurrence of P. aeruginosa varied among the sampled sub-locations, with 65% (13/20) of the isolates derived from Milimani sub-location, mainly in households (69%, 9/13) and school (31%,4/13). This observation could be attributable to the high coverage of old water plumbing systems, probably made of materials that allow for accumulation of biofilms.35 Sink/tap contamination may also be another factor,27 considering that Milimani had the highest water distribution coverage with more than 85% (6/7) of sampled households having multiple plumping fixtures (taps and sinks) as well as consistent flow of piped water. This suggests that regular cleaning and removal of biofilms from old plumbing systems such as those found in Milimani sub-location could reduce the risks of P. aeruginosa infections.
An interesting observation was that the same species of pseudomonads were isolated at different sampling points within the same household in three of the houses in Milimani, highlighting the possibility of cross-contamination at the household level as opposed to water distribution system contamination. We isolated no pseudomonad from Nyalenda A and B sub-locations, where up to 10 households used single stand-alone taps commonly located in an open space with no associated sinks or drains. We had a similar observation from households using short-term water holding and ferrying 20-litre-containers. These findings suggest the role of sinks and drains in pseudomonads transmission in communities.
In this study, we isolated P. fluorescence, P. oleovarans, P. alcaligenes, P. mendocina, P. putida and P. stutzeri in water sources at a prevalence of 7.4% (22/297). Even though not as common as P. aeruginosa, these pseudomonads were previously isolated from clinical specimens in KCRH and elsewhere, with P. stutzeri reported in wound and blood,36 P. putida in neonatal bacteremia and sepsis,37 P. fluorescens in respiratory infections,38 P. alcaligenes in bloodstream infections,19 and P. mendocina in infective endocarditis and central nervous system infections among others.20 These findings highlight the increasing clinical importance of non-P. aeruginosa pseudomonads and the need to track their environmental reservoirs. This study found these species in taps and sinks reflecting their largely environmental origin and these sources as key transmission points in the community and hospitals.
Pseudomonas species are well known for their intrinsic resistance to a wide range of antimicrobials, mainly due to the synergies between their low outer membrane permeability, possession of multidrug efflux pumps and inbuilt antimicrobial inactivation.39 In the current study, antibiotic non-susceptibility among the pseudomonads ranged from 19.5% for levofloxacin and colistin and 62% for piperacillin. Carbapenem- resistant-P. aeruginosa (CRPA) isolates, based on meropenem non-susceptibility, constituted 40% of all pseudomonads, which was consistent with 31%40 and 40%41 observed in clinical isolates (unpublished data), but lower than a recent study where CRPA were 100% non-susceptible from community and hospital acquired infections in a Kenyan referral hospital.42 The variation in these findings may be attributable to differences in the selection of study isolates, among other factors, but may suggest increased over-reliance on or misuse of carbapenem in Kisumu County, Kenya.
The prevalence of MDR pseudomonads was 45% in our study, a higher occurrence than 31% recently documented in Kenyan hospitals.40 In the current study, the highest proportion of the MDR isolates was P. aeruginosa, mainly from the community (23%, 10/24), higher than the 13.4 % reported from environmental isolates in Ghana.43 Our study found only one MDR CRPA in the hospital environment (2.3%, 1/42), which corroborates the 0.3% reported by Odoyo and others from environmental samples in Kenyan hospitals,44 highlighting the lower risk of MDR Pseudomonas infections from hospital environment reservoirs than community reservoirs. Other MDR pseudomonads, specifically P. fluorescens, were isolated from the hospital environment at a 9.5% (4/42) rate. Previous studies suggest that P. fluorescens MDR phenotypes are naturally abundant or may result from human environmental activities.45 MDR pseudomonads in the natural environment may carry mobile resistance genes which could be a reservoir for non-MDR but more pathogenic pseudomonads that, when the selection pressure from the overuse of carbapenems is applied, could drive MDR infections. All pseudomonad isolates had a MARI greater than 0.43, suggesting a high selective pressure in the current study environments, probably due to widespread antibiotic use and abuse, as reported in a previous study on the use and disposal of antibiotics in households in Kisumu, Kenya.46 To prevent dissemination of MDR-pseudomonads, protecting water systems from contamination, thus incorporating the principle of one-health where environmental health translates directly to human health remains a top priority.
This study reveals a high prevalence of MDR pseudomonads, including CRPA strains in community tanks, taps, sinks, and other water sources, presenting a critical public health threat, especially among immunocompromised persons and acting as potential conduits or reservoirs of drug resistance. Interventions, such as regular cleaning of water storage facilities, water treatment, maintenance of water piping systems, and strict implementation of antimicrobial stewardship programs, are urgently required to prevent increased resistance due to drug overuse and to eliminate environmental factors that place the vulnerable populations at risk.
The limited sampling duration and distribution and techniques may have limited the detection of pseudomonads due to temporal, diurnal and culture requirement variations. Genomic characterization of the study isolates would further identify strain types and inform of the transmission patterns of isolates based on their relatedness between the environmental sources, as well as AMR mechanisms.
Conceptualization: L.M., A.M., P.M. Funding acquisition: L.M. Investigation- P.M., AM., and E.O. Data analysis P. M, A. M, and C.K. Writing-original draft preparation: P. M and L.M. Writing-review and editing: P.M., L.M., A.M. All authors read and approved the final manuscript.
Approval to conduct this study was obtained from the Kenyatta University’s Graduate School, Kenya Medical Research institute – Scientific Ethics Review Unit (KEMRI/SERU/CCR/0190/4117 dated February 15th 2021) and the National Commission for Science, Technology and Innovation (NACOSTI/P/20/4221). Permission to collect samples from Kisumu County Hospital was sought from the hospital management. Verbal consent was obtained and it was approved by KEMRI/SERU to collect samples from households, water distribution points owners and Kisumu County Referral Hospital management
Figshare: Study samples & isolates (dataset) DOI 10.6084/m9.figshare.25284709
https://figshare.com/s/37d537fd7cb2f0519913. 47
This project contains the following underlying data
Study samples (297) and isolates (dataset in excel format)
Data are available under the terms of Creative Commons Attribution 4.0 international license (CC BY 4.0)
The authors appreciate Paul Kimani, Angela Muraya for assistance with lab procedures and Alfred Odindo, Catherine Wawira, Martin Odindo and Paul Ojore for assistance with sample collection at KCRH.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bacteriology , Anaerobic Bacteriology ,Virology ,MDR ,Candidemia , Neonatal malaria , Phytochemicals
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bacteriology, antimicrobial resistance
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 13 May 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)