Keywords
Uniprot ID P21980, TGM2, , TGM, Protein-glutamine gamma-glutamyltransferase 2, antibody characterization, antibody validation, western blot, immunoprecipitation, immunofluorescence
This article is included in the YCharOS (Antibody Characterization through Open Science) gateway.
Protein-glutamine gamma-glutamyltransferase 2 (TGM2) is a Ca2+ dependent enzyme that catalyzes transglutaminase cross-linking modifications. TGM2 is involved in various diseases, either in a protective or contributory manner, making it a crucial protein to study and determine its therapeutic potential. Identifying high-performing TGM2 antibodies would facilitate these investigations. Here we have characterized seventeen TGM2 commercial antibodies for western blot and sixteen for immunoprecipitation, and immunofluorescence. The implemented standardized experimental protocol is based on comparing read-outs in knockout cell lines against their isogenic parental controls. This study is part of a larger, collaborative initiative seeking to address antibody reproducibility issues by characterizing commercially available antibodies for human proteins and publishing the results openly as a resource for the scientific community. While the use of antibodies and protocols vary between laboratories, we encourage readers to use this report as a guide to select the most appropriate antibodies for their specific needs.
Uniprot ID P21980, TGM2, , TGM, Protein-glutamine gamma-glutamyltransferase 2, antibody characterization, antibody validation, western blot, immunoprecipitation, immunofluorescence
In version 2 of this article, we have clearly defined how the A549 WT and TGM2 KO cells were prepared and collected as lysates (medium-free) or on culture medium prior to western blot evaluation. Additionally, two columns have been added to Table 2 to indicate the immunogenic region of the antibodies as well as the number of citations for each antibody, as listed on the manufacturer catalogs and CiteAb.com, respectively.
See the authors' detailed response to the review by Michael A Rieger
See the authors' detailed response to the review by Nicoletta Bianchi
Protein-glutamine gamma glutamyltransferase 2 (TGM2) belongs to the transglutaminase family of Ca2+ dependent enzymes, regulating various cellular processes, including cell differentiation, growth and apoptosis.1–3 Encoded by TGM2 gene, TGM2 protein exhibits its function through Ca2+-dependent cross-linking of substrates, thereby modulating their activity.1 TGM2’s catalytic activity is dependent on guanine nucleotides and Ca2+ binding.4 Both GTP and/or GDP act as negative regulators of TGM2, inducing a conformational change upon binding, inhibiting its cross-linking activity (closed form). Conversely, Ca2+ binding prompts a conformational change to induce TGM2 activity (open form).5–9
Alteration and regulation of TGM2’s activity is associated with the pathogenesis of various diseases including cancer, neurodegeneration, fibrosis, inflammatory, autoimmune disorders and liver diseases.10–13 Increased TGM2 mRNA transcripts, resulting in elevated transaminase enzymatic activity, have been associated with neurodegenerative mechanism observed in Parkinson’s disease, Alzheimer’s disease and Huntington’s disease.14,15 Given that α-synuclein serves as a common substrate for TGM2, elevated activity of TGM2 results in the formation of soluble aggregates as well as insoluble inclusions; distinctive features of Parkinson’s disease pathogenesis.15–18 Regulating TGM2 activity using inhibitors could positively affect the human diseases in which TGM2 is implicated.19,20 Identifying high-quality antibodies would accelerate TGM2 research and its potential as a pharmacological target.
This research is part of a broader collaborative initiative in which academics, funders and commercial antibody manufacturers are working together to address antibody reproducibility issues by characterizing commercial antibodies for human proteins using standardized protocols, and openly sharing the data.21–23 Here, we evaluated the performance of seventeen commercially-available antibodies for TGM2 for use in western blot, and sixteen for immunoprecipitation and immunofluorescence, enabling biochemical and cellular assessment of TGM2 properties and function. The platform for antibody characterization used to carry out this study was approved by a committee of industry and academic researchers, whose members are mentioned in the competing interest section. It consists of first identifying appropriate human cell lines, development/contribution of equivalent knockout cell lines and finally following antibody characterization procedures on commonly used commercial antibodies. The standardized consensus antibody characterization protocols are openly available on Protocol Exchange, DOI: 10.21203/rs.3.pex-2607/v1.24
Our standard protocol involves comparing readouts from wild-type (WT) and knockout (KO) cells.25,26 The first step is to identify a cell line(s) that expresses sufficient levels of a given protein to generate a measurable signal. To this end, we examined the DepMap transcriptomics database to identify all cell lines that express the target at levels greater than 2.5 log2 (transcripts per million “TPM” + 1), which we have found to be a suitable cut-off (Cancer Dependency Map Portal, RRID:SCR_017655). Commercially available A549 cells expressed the TGM2 transcript at RNA levels above the average range of cancer cells analyzed. Parental and TGM2 KO A549 cells were obtained from Abcam (Table 1).
| Institution | Catalog number | RRID (Cellosaurus) | Cell line | Species of origin | Genotype |
|---|---|---|---|---|---|
| Abcam | ab275463 | CVCL_0023 | A549 | Homo sapiens (Human) | WT |
| Abcam | ab261876 | CVCL_B1I0 | A549 | Homo sapiens (Human) | TGM2 KO |
For western blot experiments, WT and TGM2 KO cells are collected as lysates or with a serum-free medium, ran on SDS-page, transferred on nitrocellulose membranes, and then probed with all antibodies in parallel (Figures 1 and 2).
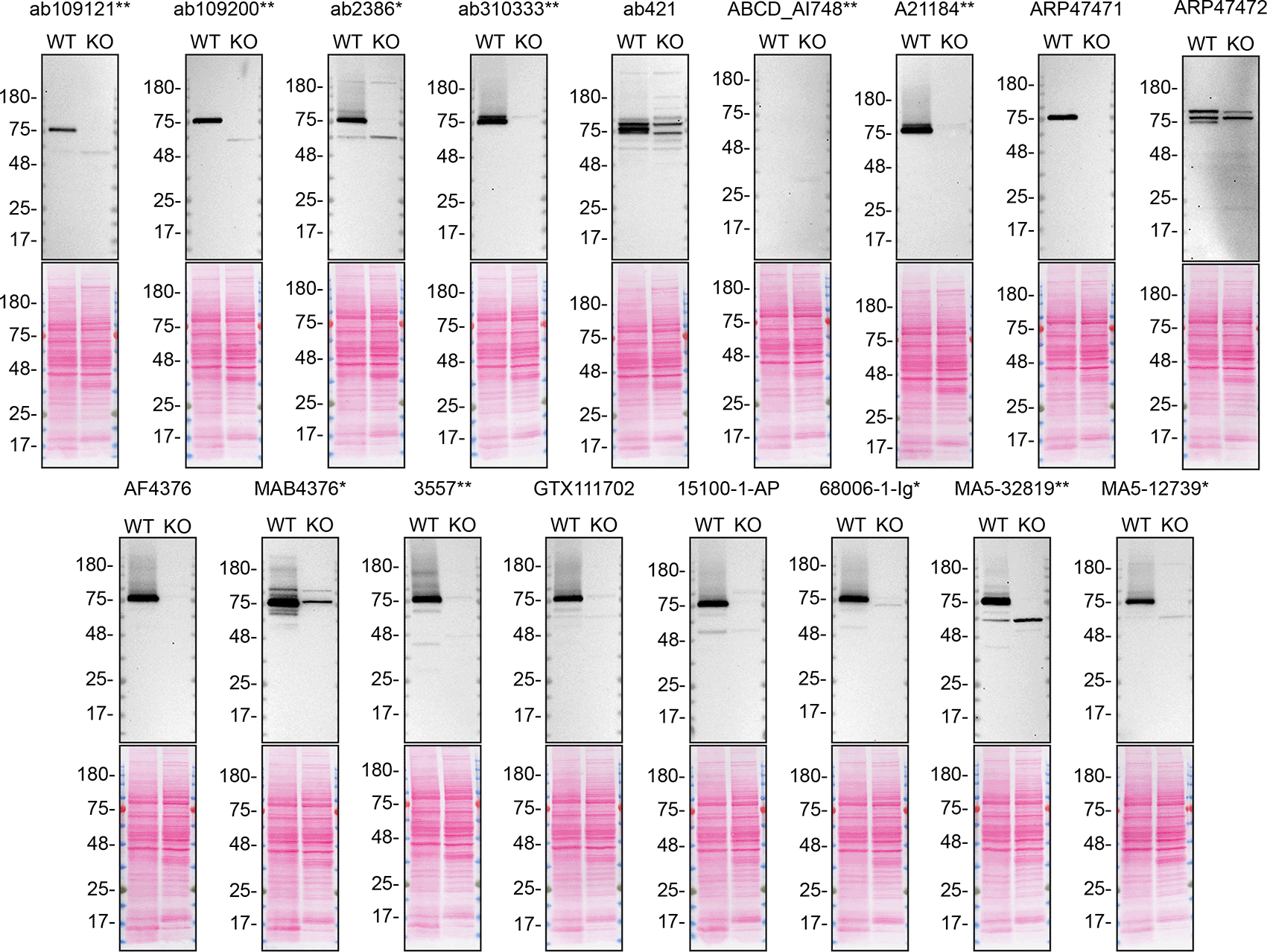
Lysates of A549 (WT and TGM2 KO) were prepared and 40 μg of protein were processed for western blot with the indicated TGM2 antibodies. The Ponceau stained transfers of each blot are presented to show equal loading of WT and KO lysates and protein transfer efficiency from the acrylamide gels to the nitrocellulose membrane. Antibody dilutions were chosen according to the recommendations of the antibody supplier. Antibody dilution used: ab109121** at 1/1000, ab109200** at 1/10000, ab2386* at 1/500, ab310333** at 1/1000, ab421 at 1/500, ABCD_AI748** at 1/10, A21184** at 1/10000, ARP47471 at 1/500, ARP47472 at 1/500, AF4376 at 1/400, MAB4376* at 1/200, 3557** at 1/500, GTX111702 at 1/500, 15100-1-AP at 1/6000, 68006-1-Ig* at 1/10000, MA5-32819** at 1/500, MA5-12739* at 1/200. Predicted band size: 77 kDa. *Monoclonal antibody, **Recombinant antibody.
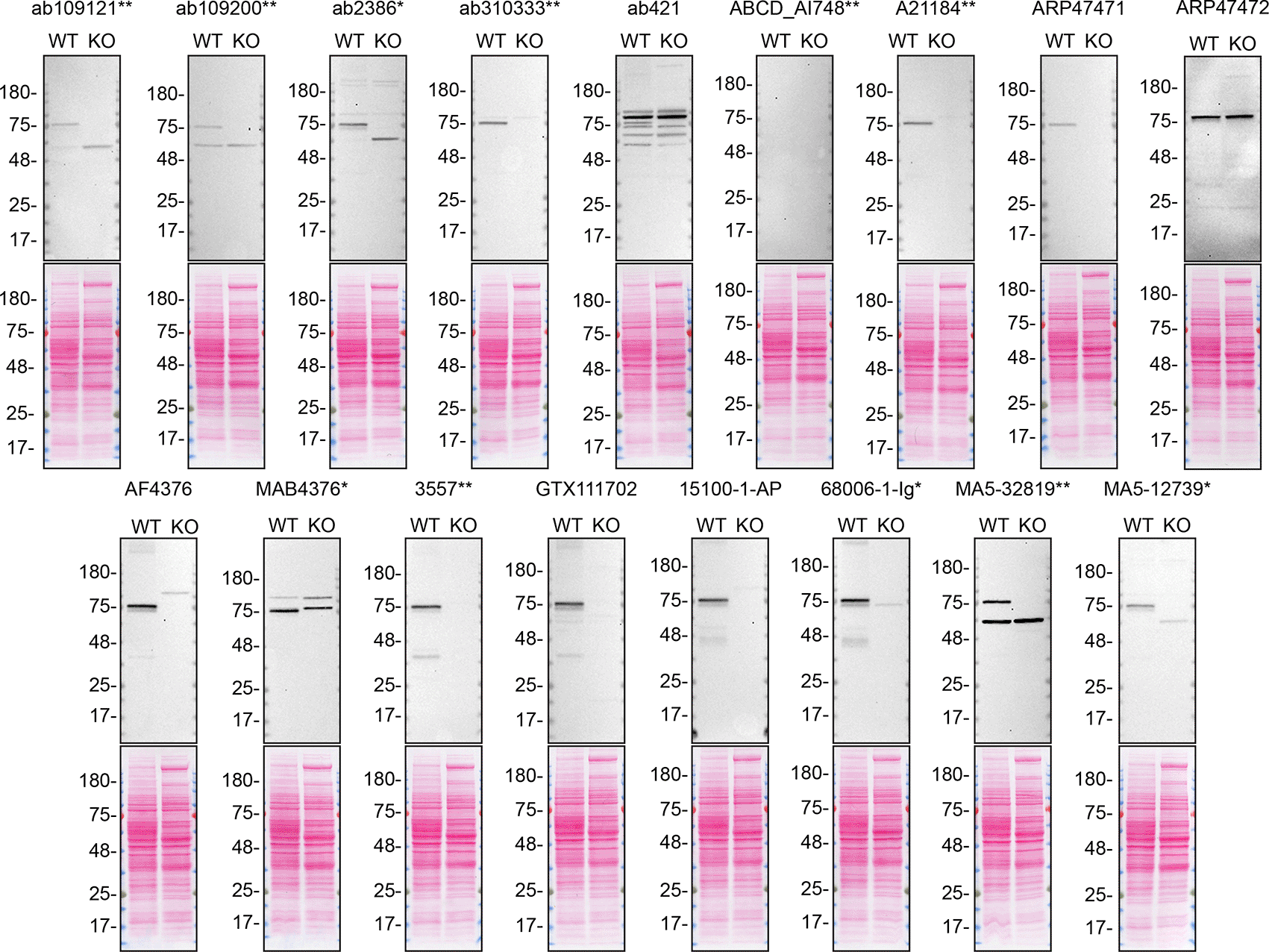
A549 WT and TGM2 KO were cultured in serum free media, and 40 μg of protein from concentrated culture media were processed for western blot with the indicated TGM2 antibodies. The Ponceau stained transfers of each blot are shown. Antibody dilution used: ab109121** at 1/1000, ab109200** at 1/10000, ab2386* at 1/500, ab310333** at 1/1000, ab421 at 1/500, ABCD_AI748** at 1/10, A21184** at 1/10000, ARP47471 at 1/500, ARP47472 at 1/500, AF4376 at 1/400, MAB4376* at 1/200, 3557** at 1/500, GTX111702 at 1/500, 15100-1-AP at 1/6000, 68006-1-Ig* at 1/10000, MA5-32819** at 1/500, MA5-12739* at 1/200. Predicted band size: 77 kDa. *Monoclonal antibody, **Recombinant antibody.
As per our standard procedure, we next used the antibodies to immunoprecipitate TGM2 from A549 cell extracts. To evaluate the performance of each antibody, the TGM2 protein was detected in extracts, in each extract unbound to the antibody and corresponding immunoprecipitates (IP) (Figure 3). To detect TGM2, a western blot was performed with an antibody successful under the conditions tested in Figure 1.
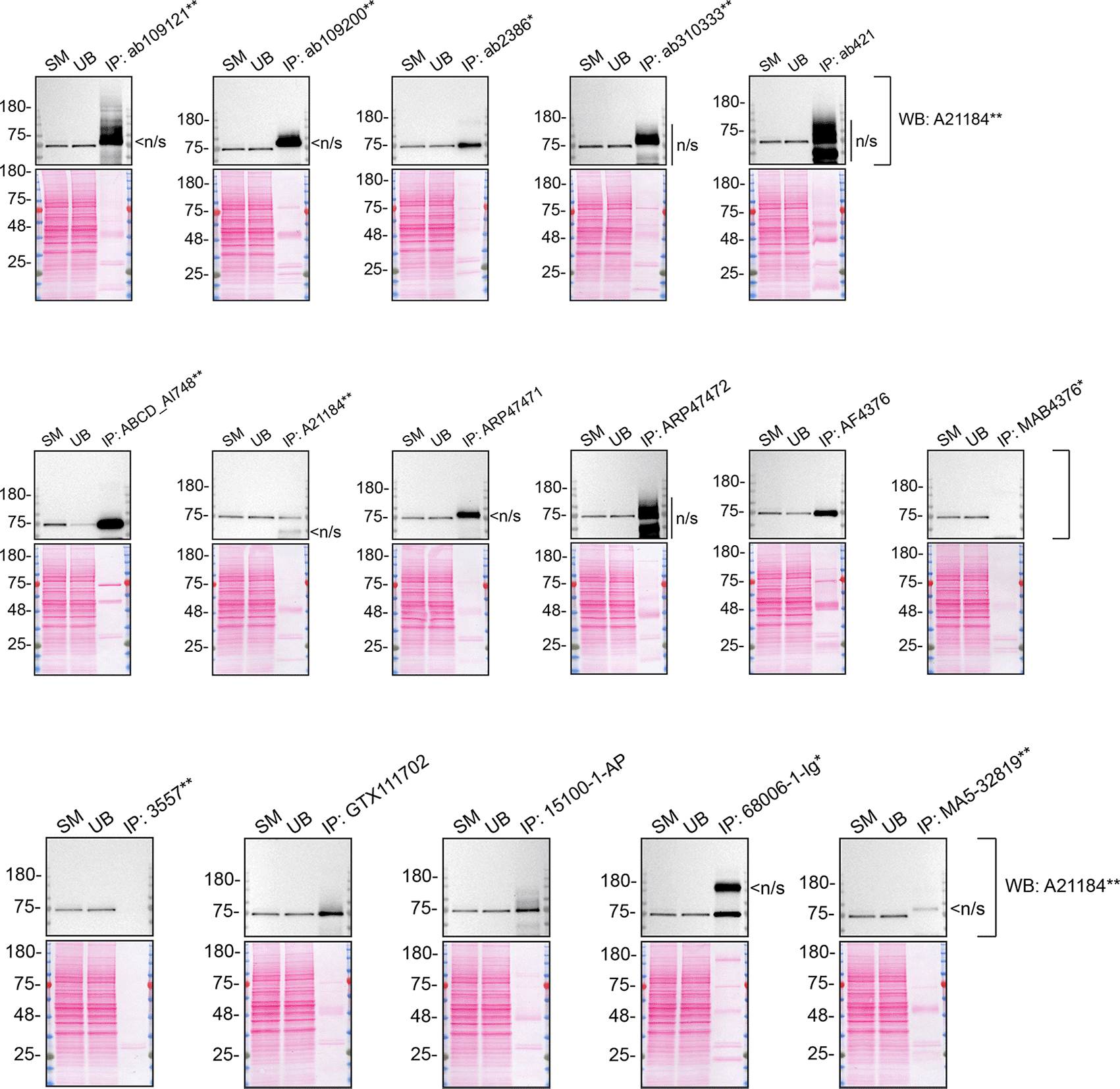
A549 lysates were prepared, and immunoprecipitation was performed using 2.0 μg of the indicated TGM2 antibodies pre-coupled to Dynabeads protein A or protein. Samples were washed and processed for western blot with the indicated TGM2 antibody. For western blot, A21184** was used at 1/10000. The Ponceau stained transfers of each blot are shown. SM=4% starting material; UB=4% unbound fraction; IP=immunoprecipitate; n/s=non-specific signal. *Monoclonal antibody, **Recombinant antibody.
For immunofluorescence, antibodies were screened using a mosaic strategy, as per our standard procedure. First, A549 WT and TGM2 KO were labelled with different fluorescent dyes in order to distinguish the two cell lines, and TGM2 antibodies were evaluated. Cells were imaged in the same field of view to reduce staining, imaging and image analysis bias (Figure 4). Quantification of immunofluorescence intensity in hundreds of WT and KO cells was performed for each antibody tested. The images presented in Figure 4 are representative of the results of this analysis.
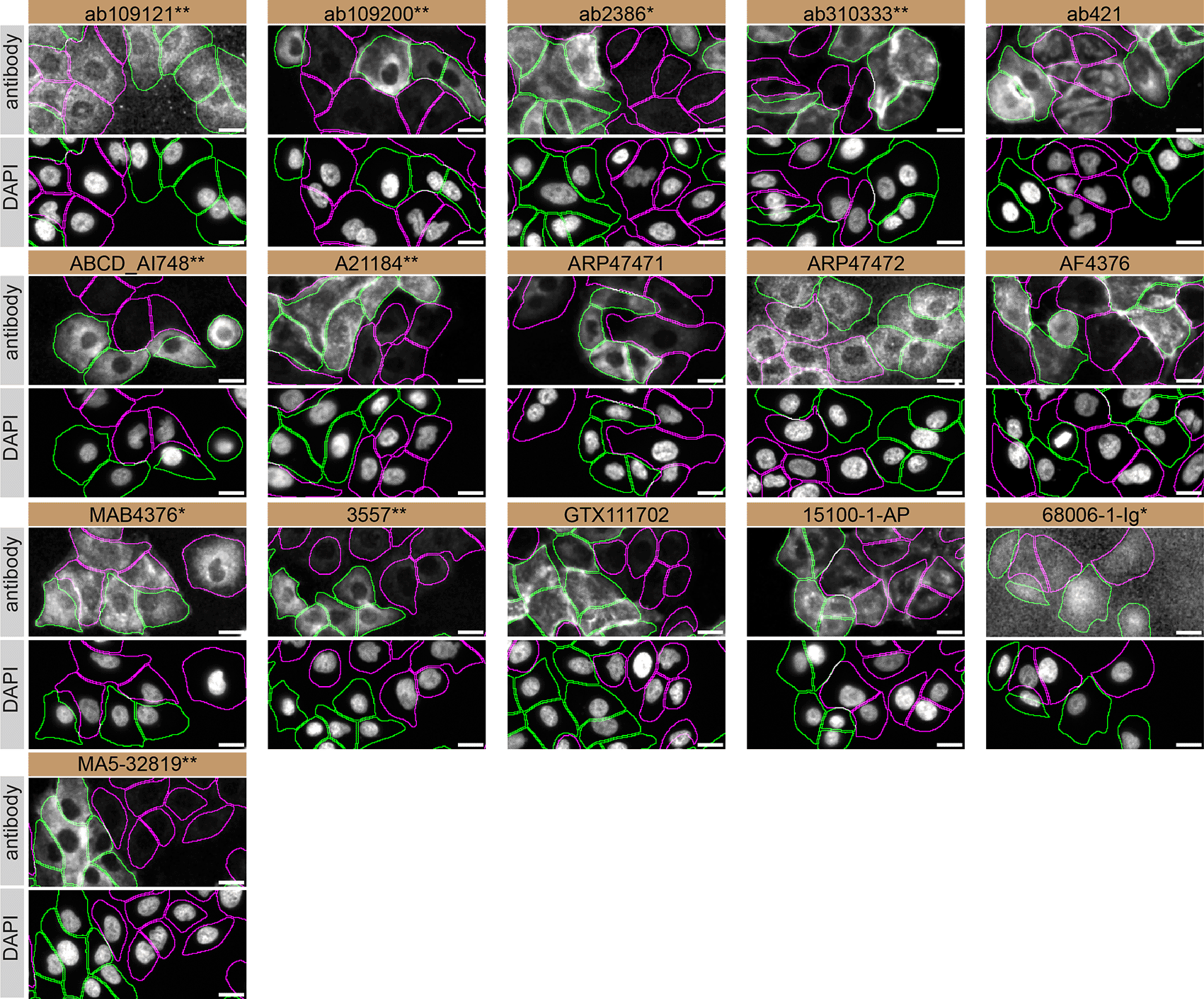
A549 WT and TGM2 KO cells were labelled with a green or a deep-red fluorescent dye, respectively. WT and KO cells were mixed and plated to a 1:1 ratio in a 96-well plate with optically clear flat-bottom. Cells were stained with the indicated TGM2 antibodies and with the corresponding Alexa-fluor 555 coupled secondary antibody including DAPI. Acquisition of the blue (nucleus-DAPI), green (identification of WT cells), red (antibody staining) and deep-red (identification of KO cells) channels was performed. Representative images of the merged blue and red (grayscale) channels are shown. WT and KO cells are outlined with green and magenta dashed line, respectively. When the concentration was not indicated by the supplier, which was the case for all antibodies tested, except ab310333**, ABCD_AI748 and A21184**, we tested antibodies at using the dilutions listed below. At these concentrations, the signal from each antibody was in the range of detection of the microscope used. Antibody dilution used: ab109121** at 1/1000, ab109200** at 1/300, ab2386* at 1/1000, ab310333** at 1/500, ab421 at 1/1000, ABCD_AI748** at 1/1000, A21184** at 1/1000, ARP47471 at 1/500, ARP47472 at 1/250, AF4376 at 1/100, MAB4376* at 1/100, 3557** at 1/500, GTX111702 at 1/1000, 15100-1-AP at 1/300, 68006-1-Ig* at 1/500, MA5-32819** at 1/1000. Bars = 10 μm. *Monoclonal antibody, **Recombinant antibody.
In conclusion, we have screened seventeen TGM2 commercial antibodies by western blot, and sixteen by immunoprecipitation, and immunofluorescence, comparing the signal produced by the antibodies in human A549 WT and TGM2 KO cells. Several high-quality antibodies that successfully detect TGM2 under our standardized experimental protocol can be identified. Researchers who wish to study TGM2 in a different species are encouraged to select high-quality antibodies, based on the results of this study, and investigate the predicted species reactivity of the manufacturer before extending their research.
In our effort to address the antibody reliability and reproducibility challenges in scientific research, the authors recommend the antibodies that demonstrated to be underperforming under our standard procedure be removed from the commercial antibody market. Following the release of the antibody characterization, ab421 was removed from the manufacturer’s antibody catalog.
The authors do not engage in result analysis or offer explicit antibody recommendations. A limitation of this study is the use of universal protocols - any conclusions remain relevant within the confines of the experimental setup and cell line used in this study. Our primary aim is to deliver top-tier data to the scientific community, grounded in Open Science principles. This empowers experts to interpret the characterization data independently, enabling them to make informed choices regarding the most suitable antibodies for their specific experimental needs. Guidelines on how to interpret antibody characterization data found in this study are featured on the YCharOS gateway.27
The underlying data for this study can be found on Zenodo, an open-access repository for which YCharOS has its own collection of antibody characterization reports.28,29
The standardized protocols used to carry out this KO cell line-based antibody characterization platform was established and approved by a collaborative group of academics, industry researchers and antibody manufacturers. The detailed materials and step-by-step protocols used to characterize antibodies in western blot, immunoprecipitation and immunofluorescence are openly available on Protocol Exchange, a repository dedicated to openly sharing scientific research protocols, DOI: 10.21203/rs.3.pex-2607/v1.24
Cell lines used and primary antibodies tested in this study are listed in Table 1 and 2, respectively. To ensure that the cell lines and antibodies are cited properly and can be easily identified, we have included their corresponding Research Resource Identifiers, or RRID.30,31
| Company | Catalog number | Lot number | RRID (Antibody Registry) | Clonality | Clone ID | Host | Immunogenic region | Concentration (μg/μl) | Vendors recommended applications | Number of citations (CiteAb.com) |
|---|---|---|---|---|---|---|---|---|---|---|
| Abcam | ab109121** | 1058833-3 | AB_10861115 | Recombinant-mono | EPR2956 | rabbit | n/a | 2.79 | Wb | 1 |
| Abcam | ab109200** | 1044092-1 | AB_10860177 | Recombinant-mono | EP2957 | rabbit | proprietary information | 0.30 | Wb | 12 |
| Abcam | ab2386* | 1051063-6 | AB_2287299 | Monoclonal | CUB 7402 | mouse | proprietary information | n/a | Wb, IF | 49 |
| Abcam | ab310333** | 1056093-5 | AB_3076417 | Recombinant-mono | EPR28142-86 | rabbit | proprietary information | 0.50 | Wb, IF | 0 |
| Abcam | ab421 | 1034725-7 | AB_304372 | Polyclonal | - | rabbit | proprietary information | n/a | Wb, IF | 37 |
| ABCD | ABCD_AI748** | 10/27/2023 | AB_3076341 | Recombinant-mono | 679-14-E06 | rabbit | proprietary information | 0.12 | Others | 0 |
| ABclonal | A21184** | 3522042510 | AB_3083448 | Recombinant-mono | ARC52843 | rabbit | aa 438-687 | 1.30 | Wb, IF | n/a |
| Aviva Systems Biology | ARP47471 | QC18320-43546 | AB_1107120 | Polyclonal | - | rabbit | middle region | 0.50 | Wb | n/a |
| Aviva Systems Biology | ARP47472 | QC16720 | AB_1088480 | Polyclonal | - | rabbit | N-terminal | 0.50 | Wb | n/a |
| R&D Systems (a Bio-Techne brand) | AF4376 | CFGU0119031 | AB_10890213 | Polyclonal | - | sheep | recombinant fragment (Ala2-Ala687) | 0.20 | Wb | 4 |
| R&D Systems (a Bio-Techne brand) | MAB4376* | CFNO0119031 | AB_10971763 | Monoclonal | 716620 | mouse | recombinant fragment (Ala2-Ala687) | 0.20 | Wb | 2 |
| Cell Signaling Technology | 3557** | 3 | AB_2202883 | Recombinant-mono | D11A6 | rabbit | proprietary information | 0.02 | Wb | 41 |
| GeneTex | GTX111702 | 44524 | AB_1952227 | Polyclonal | - | rabbit | middle region | 1.05 | Wb, IF | 8 |
| Proteintech | 15100-1-AP | 00081307 | AB_2202885 | Polyclonal | - | rabbit | recombinant fragment (Ag7439) | 0.60 | Wb, IP, IF | 21 |
| Proteintech | 68006-1-Ig* | 10023724 | AB_2918753 | Monoclonal | 2D4C11 | mouse | recombinant fragment (Ag7462) | 1.00 | Wb, IF | n/a |
| Thermo Fisher Scientific | MA5-32819** | YJ4089240 | AB_2810095 | Recombinant-mono | JU30-02 | rabbit | aa 578-627 | 1.00 | Wb | 0 |
| Thermo Fisher Scientific | MA5-12739* | ZA4176225 | AB_10985077 | Monoclonal | CUB 7402 | mouse | Purified guinea pig liver TGase II | 0.20 | Wb, IP, IF | 115 |
Zenodo: Antibody Characterization Report for TGM2 (Protein-glutamine gamma-glutamyltransferase 2), https://doi.org/10.5281/zenodo.10819348. 28
Zenodo: Dataset for the TGM2 (Protein-glutamine gamma-glutamyltransferase 2) antibody screening study, https://doi.org/10.5281/zenodo.10927535. 29
Protocol Exchange: A consensus for antibody characterization platform, https://doi.org/10.21203/rs.3.pex-2607/v1. 24
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
We would like to thank the NeuroSGC/YCharOS/EDDU collaborative group for their important contribution to the creation of an open scientific ecosystem of antibody manufacturers and knockout cell line suppliers, for the development of community-agreed protocols, and for their shared ideas, resources and collaboration. Members of the group can be found below. We would also like to thank the Advanced BioImaging Facility (ABIF) consortium for their image analysis pipeline development and conduction (RRID:SCR_017697). Members of each group can be found below.
NeuroSGC/YCharOS/EDDU collaborative group: Thomas M. Durcan, Aled M. Edwards, Peter S. McPherson, Chetan Raina and Wolfgang Reintsch.
ABIF consortium: Claire M. Brown and Joel Ryan.
Thank you to the Structural Genomics Consortium, a registered charity (no. 1097737), for your support on this project. The Structural Genomics Consortium receives funding from Bayer AG, Boehringer Ingelheim, Bristol-Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute (grant no. OGI-196), the EU and EFPIA through the Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant no. 875510), Janssen, Merck KGaA (also known as EMD in Canada and the United States), Pfizer and Takeda.
An earlier version of this of this article can be found on Zenodo (DOI: 10.5281/zenodo.10819348).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Membrane trafficking, RAB GTPases, phosphoinositides, Drosophila, proteomics, autophagy.
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: cancer research
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: cancer research
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Stem cells, Cancer stem cells, Single cell technologies, TGM2 physiology in colorectal cancer and other cancers
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 30 Jul 24 |
read | read | |
|
Version 1 17 May 24 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)