Keywords
Cassava, Seedling nursery stage, Clonal evaluation trial, Inbreeding depression, S1 progenies, Heterozygosity index, SNPs markers, Cassava improvement
This article is included in the Plant Science gateway.
Cassava is an outcrossing, highly heterozygous plant that is reported to suffer from inbreeding depression. However, unraveling recessive traits and exploring additive genes would require a limited level of inbreeding for the genetic improvement of cassava.
The impact of inbreeding depression (ID) on agronomic and biotic tolerance of cassava was evaluated using the S1 progenies of five African cassava varieties (TMS 30572, TME 419, TMS 98/0505, TMS 01/1371, and TMS 98/0002) at the seedling and clonal evaluation stages.
At both trial stages, the effects of ID were severe on average performance of fresh root yield and fresh foliage yield; moderate on harvest index, dry matter content, vigor, and tolerance to cassava mosaic disease, cassava bacterial blight, and cassava anthracnose diseases; and less severe on plant height. Further examination of S1 families using molecular markers showed varying levels of heterozygosity at several loci across the genome. In addition, the degree of heterozygosity and ID varied by the S1 family. The TMS 01/1371 family showed the lowest degree of heterozygosity and ID on the average performance of different agronomic attributes, indicating that inbreeding may be strategically explored in this family to increase genetic gain and identify useful recessive traits. Lastly, the observed depression from inbreeding was higher in seedling evaluation than in clonal evaluation trials for fresh root yield, fresh foliage yield, harvest index, and dry matter content.
S1 individuals showing relatively low heterozygosity based on SNP data were selected as parents to advance genetic gain in cassava. Generally, reduced heterozygosity was prominent in traits with severe ID impacts on average phenotypic performance. Our results highlight the relative importance of exploring non-additive genetic effects and transgressive segregations for favorable allele combinations in cassava improvement.
Cassava, Seedling nursery stage, Clonal evaluation trial, Inbreeding depression, S1 progenies, Heterozygosity index, SNPs markers, Cassava improvement
Cassava (Manihot esculenta Crantz) plays a vital role as one of the primary sources of energy in the diets of most tropical countries. Additionally, its significance has grown, as it now serves as a crucial raw material for diverse industries. Consequently, understanding its genetics and factors influencing trait inheritance and expression is of utmost importance for its continued development.
Inbreeding occurs when two genetically related individuals mate, increasing the likelihood of their offspring having recessive or deleterious traits. This often results in a reduced fitness of a population, known as inbreeding depression. Cassava is highly heterozygous and typical cross-pollinated species, has been associated with severe inbreeding depression (Rojas et al., 2009). Nonetheless, it is encouraging to observe that interest in inbreeding is beginning to gain momentum (Ceballos et al., 2004, 2007; Rojas et al., 2009) in cassava improvement initiatives. Certainly, there is the potential to harness the benefits of inbreeding, similar to what has been achieved with maize, by capitalizing on both additive and non-additive effects (Rojas et al., 2009). Inbreeding will provide several advantages in cassava such as elimination of harmful recessive genes. Through selection, the less desirable genes are eliminated and superior genes are begin accumulated resulting in an increase in the productivity of the inbred populations (Ceballos et al., 2004). A key objective of this study was to explore the means of efficiently utilizing additive genetic variance in trait development through the fixation of beneficial alleles for key economic traits. By fixing useful alleles, we aimed to create valuable genetic stocks that can serve as parent lines, thereby reducing segregation issues that typically arise when evaluating F1 populations in breeding schemes for the development of improved varieties in Africa. This report presents the findings of our investigations for quantifying inbreeding depression in commercial elite African cassava lines. With these insights, we hope to pave the way for leveraging inbreeding as a strategic tool to enhance cassava breeding programs and to unlock its full potential as a vital crop for sustainable development.
The field experiment was conducted at the National Root Crops Research Institute, (NRCRI) Umudike, Nigeria, while genotyping was conducted at the biotechnology laboratory of KBiosciences, Hoddesdon, London, United Kingdom. The NRCRI, Umudike, is located at latitude 5°30’N and longitude 7°30’E. It is at an altitude of approximately 122m above sea level and has an annual rainfall of approximately 2,166.3 mm, a temperature range of 22°C - 32°C, and a relative humidity range of 80–90%.
• Seed production
Five of the elite cassava genotypes (TMS 30572, TME 419, TMS 98/0505, TMS 01/1371 and TMS 98/0002) were obtained from the germplasm of International Institute of Tropical Agriculture, (IITA) Ibadan. These five elite cassava genotypes were used as progenitors (S0) to generate S1 progenies. They were not randomly selected because they were preferred for their wide adoption by farmers for favorable morphological and agronomic attributes in the elite germplasm. The elite genotypes were planted at a distance of 1 m × 1 m using 20 stem cuttings per genotype. For only self-pollination to occur, the plots were isolated by 100m apart from the neighbouring cassava plots. Self-pollination was performed for each elite clone. The pollinated flowers were secured within pollination bags in order to safeguard them from external pollen carried by bees. The bags were then carefully removed the following day. The mature fruits were harvested at 90 days after pollination, placed in a well-labelled pollination bags, and are allowed to dehiscent.
• Establishment of the seedling evaluation trial
Harvested S1 botanical seeds were allowed a two - month dormancy period before being sowed in nurseries in a screen house. The seeds were sown in trays filled with sterilized soil with a mixture of loamy and sandy soil at a ratio of 2:1 in the screen house. Seeds germinated quickly under optimal soil temperatures (30°C–35°C) and moisture regimes. They were irrigated twice daily, in the morning and evening. The seeds started germinating 10 to 12 days after planting and were transplanted when they attained a height of 15–20 cm. After two months in the nursery, S1 seedlings were transplanted to a well-prepared field where they were grown and evaluated. Harvesting was performed 12 months after planting, after which they were cloned to generate at least 10 stem cuttings per seedling for clonal evaluation.
• Establishment of the clonal evaluation trial
In the clonal evaluation trial, ten cuttings of each S1 genotype were planted in the field for evaluation. All progenies belonging to the same family, together with the parents, were established in the same block in an augmented (Federer, 1956) experimental design. In this design, each clone or genotype was planted in a row and are not replicated while the parents serve as checks and are replicated in each block. The spacing within rows was 1 m, and the row spacing was 1.5 m to minimize inter-plot interference.
Genomic DNA was extracted from the leaf samples of each individual of the S1 population (from five families) and from the parents, as described by Dellarporta et al. (1983). A total of 200 SNPs markers with good genome distribution and coverage were analyzed in 260 DNA samples. Constituent reagent volumes for the KBioscience Competitive Allele-specific PCR genotyping system (KASP) genotyping mix were as follows:
• 5 μl DNA
• 5 μl KASP 2X reaction mix
• 12 μl of Allele-specific primer 1 (100 μM)
• 12 μl of Allele-specific primer 2 (100 μM)
• 30 μl of common (reverse) Primer (100 μM)
• 46 μl of H2O/Tris-HCl (10 mM, pH 8.3)
Procedure
5 μl DNA samples were arrayed on a PCR plate. The DNA was genotyped as a liquid sample. The KASP genotyping mix was dispensed robotically into PCR plates containing 5 μl DNA samples and briefly vortexed. The plate was sealed with an optically clear seal and thermally cycled using a Peltier block-based thermocycler at 94°C for 15 minutes. The plate was read using a suitable fluorescent plate reader at ambient temperature. Genotyping data were analyzed using the most FRET-capable plate readers.
During the growth period of the seedling nursery and clonal trials, the plants were evaluated for Cassava Mosaic Disease (CMD), cassava bacterial light (CBB), Cassava Anthracnose Disease (CAD), and vigor 1, 3, 6, 9, and 12 Months After Planting (MAP).
A. Vigor was evaluated based on a scale of 1-5; where 1 = very weak (plants somewhat stunted with very thin stems), 2 = weak (thin stem), 3 = intermediate (moderately growing), 4 = vigorous (fast growing and no bending), and 5 = extra vigorous (very fast growing, strong, and no bending) (IITA, 1990).
B. Cassava Mosaic Disease (CMD) was visually scored on a scale of 1-5; where 1 = no symptoms observed; 2 = mild chlorotic pattern on the entire leaflet; 3 = strong mosaic pattern on the entire leaf; 4 = severe mosaic/distortion of 2/3 of leaflet; and 5 = severe mosaic/distortion of 4/5 of leaflet.
C. Cassava Bacterial Blight (CBB) was visually scored on a scale of 1-5; where 1 = no symptoms observed; 2 = only angular leaf spots; 3 = exclusive leaf blight, leaf wilt and defoliation, and gum exudation on the stem; 4 = exclusive leaf blight, defoliation, and die-back; and 5 = complete defoliation and stem die-back.
D. Cassava Anthracnose Disease (CAD) was visually characterized. A scale of 1 to 5 was used to rate symptoms: 1 = no symptoms observed; 2 = few shallow cankers on woody stems; 3 = many deep cankers on woody stems; 4 = many oval lesions on green stems; and 5 = many lesions on green stems and severe necrosis at leaf axils.
At harvest for seedling and clonal evaluation trials at 12 MAP, plant height (cm) was measured from the soil level to the highest apical point of the plant at harvest. Phenotypic assessments were performed on eight innermost plants per clone. Roots were separated from the vegetative harvestable biomass (leaves, stems, and the original planting stake) and independently weighed using a weighing balance. The fresh root yield and fresh foliage yield (kg plant−1) were calculated. The harvest index (root biomass as a proportion of total biomass) was computed for each clone following the procedure outlined by Kawano (1990). Estimation of Dry Matter Content (DMC) (measured as a percentage) in the root samples was determined using the specific gravity method, as suggested by Kawano et al. (1987). The procedure included weighing of three (3) kg of roots with a weighing balance and then placing them in a nylon bag. The bag with roots was hung on a hanging balance of 6 kg capacity while suspending the bag in a tank with water, ensuring that the bag did not touch the bottom and that the roots were completely underwater. The weight of the water was then determined.(IITA, 1990)
The number of progenies assessed differed across the families, making the dataset largely uneven. Summary statistics (average, range, variance, and skewness) for all traits were calculated for each family. The dataset for all the traits was subjected to a paired sample t-test using R. Inbreeding depression was estimated for plant height, vigor, fresh root yield, fresh foliage yield, harvest index, dry matter content, and disease data; cassava mosaic disease, cassava bacterial blight, and cassava anthracnose disease as a percentage of the S1 average. ID was obtained based on the equation reported by Rojas et al. (2009):
Inbreeding depression was assessed at the family level. The lower the ID value, the more severe the level of depression, indicating that the performance of the S1 progenies is relatively close to that of the S0 progenitor compared to the ID values that have higher depression.
The heterozygosity index was obtained using the following equation suggested by Scotti et al. (2000):
Where Sn is the inbreeding generation in which the effect was calculated, Sn-x is the generation taken as a reference (the parental S0), and H is the mean of heterozygosity loci. Data were analyzed using GenStat version 12.1 edition (2009).
Table 1 presents the origin of the elite clones and the sizes of their respective S1 families, which were evaluated in both seedling nursery and clonal evaluation trials. The selection of these families (TME 419, TMS 01/1371, TMS 30572, TMS 98/0002 and TMS 98/0505) for evaluation was based on the commercial significance of their progenitors in Africa. Specifically, family TME 419 started with 51 seeds, of which 33 genotypes were evaluated in seedling nurseries and 23 were evaluated in clonal evaluation trials. Similarly, for family TMS 01/1371, 79 seeds were produced, 63 were evaluated in the seedling nursery stage, and 35 were evaluated in the clonal stage. In the case of family TMS 30572, out of 111 seeds produced, 90 genotypes survived the seedling stage, and after the usual losses in the cloning process, 51 clones were evaluated in the clonal stage. For family TMS 98/0002, 44 out of the initial 64 seeds were evaluated in the seedling nursery and 24 in the clonal stage. Lastly, family TMS 98/0505 began with 68 seeds, but only 25 cloned genotypes were evaluated at the seedling stage, and only 14 cloned genotypes were evaluated at the clonal stage. These losses are typical and are also observed in the evaluation of the full-vigor germplasm. Despite these losses, the samples of S1 genotypes representing the partially inbred populations were considered unbiased representations of all progenies that could be obtained from the self-pollination of the respective progenitors. This selection process ensures a fair representation of the evaluated progenies and allows for a comprehensive assessment of the performance and potential of partially inbred populations in breeding programs.
Inbreeding depression (ID) is generally lower for pests and diseases than for complex traits such as yield. Inbreeding depression as a percentage of the performance from the S0 generation measured in five S1 families based on disease severity in seedling and clonal evaluation trials are presented in Tables 2 and 3, respectively. The disease data had negligible additive effects and had low ID values.
In the seedling evaluation trial, CMD had the lowest average ID value (-94.30%), which ranged from -141.00% (TME 419) to 0.50% (TMS 30572), followed by CBB with an average ID value of -71.90% and with a range from -175.00% (TMS 98/0002) to 7.00% (TMS 30572), followed by CAD with an average ID value of -46.80%, ranging from -117.00% (TMS 98/0505) to 23.00% (TMS 30572) (Table 2). In the clonal evaluation trial, CMD had the lowest average ID value (-41.08%), ranging from -57.80% (TMS 30572) to -16.57% (TMS 01/1371), followed by CAD with an average ID value of -33.44% and ranging from -54.90% (TMS 30572), followed by CBB with an average ID value of -21.66%, ranging from -46.83% (TMS 98/0505) to 9.14% (TMS 01/1371) (Table 3).
Phenotypic assessment of the families revealed some superior S1 clones, which were found to substantially outperform their S0 progenitors. In the case of cassava mosaic disease, cassava bacterial blight, and cassava anthracnose disease, no significant depression was observed (Tables 2 and 3).
Agronomic traits revealed notable ID, with fresh root yield exhibiting the highest ID values at 86.55% and 53.05% at the seedling nursery and clonal evaluation stages, respectively (Tables 4 and 5). Other agronomic traits, such as fresh foliage yield and harvest index, also displayed considerable ID values (38% and 22%, respectively) at the clonal evaluation stage (Table 5). The effects of ID tended to be relatively higher for more complex traits given their polygenic character. Reduced heterozygosity associated with selfing (S1) had a significant impact on fresh root yield and foliage, underscoring the relative importance of non-additive genetic effects in their inheritance. Some transgressive segregants were observed in the S1.
As with pests and diseases, ID was lower than that of the complex traits. The average ID for vigour in the seedling evaluation trial was 20.83% ranging from -3.03% in family TMS 01/1371 to 44.44% in family TMS 30572 (Table 4) while in clonal evaluation trial, the average ID value for vigour was 28.22% ranging from 1.71% in family TMS 01/1371 to 46.47% in family TMS 30572 (Table 5). For plant height, the average ID observed at the seedling nursery and clonal evaluation stages were -25.49% and -6.50%, respectively, and all families produced S1 genotypes with plant heights superior to those of their S0 progenitor genotypes in both stages. This demonstrated that the performance of the progenies was relatively close to that of the S0 progenitors. Based on these results, this set of traits appears to be less sensitive to inbreeding depression.
A paired (dependent) sample t-test was used to compare the means of the seedling nursery and clonal evaluation stages to determine whether there was a statistically significant difference between these means. The results revealed a significant difference between the seedling nursery and clonal evaluation stages for all traits evaluated (Table 6). There was a significant difference in the CMD scores for the seedling nursery (M = 1.56, SD = 0.62) and clonal evaluation trials (M = 2.05, SD = 0.79); t (146) = 5.74, p = 0.000. For CBB, the clonal evaluation trial showed a significantly higher level of resistance (M = 2.44, SD = 0.39) than the seedling nursery stage (M = 3.01, SD = 0.70), t (146) = 8.25, p = 0.000. Similarly, the mean score for CAD at the clonal evaluation stage (M = 1.54, SD = 0.2) showed a higher level of resistance than the mean score at the seedling nursery stage (M = 2.26, SD = 0.54); t (146) = 15.72, p = 0.000.
More vigorous plants were observed at the clonal evaluation stage (M = 3.23, SD = 0.64) than at the seedling nursery stage (M = 2.52, SD = 0.95; t (146) = 7.92, p = 0.000). There was a significant difference between the heights of plants at the seedling nursery (M = 174.22, SD = 50.16) and clonal evaluation stages (M = 204.44, SD = 57.06); t (146) = -4.87, p = 0.000.
There was a significant improvement in fresh foliage yield from the seedling nursery stage (M = 1.44, SD = 1.81) to the clonal evaluation stage (M = 5.17, SD = 5.03); t (146) = -8.42, p = 0.000. The result also showed that fresh root yield was significantly higher at the clonal evaluation stage (M = 17.28, SD = 13.22) than at the seedling nursery stage (M = 1.53, SD = 1.78); t (146) = -3.72, p = 0.000. The dependent sample t-test showed that the seedling nursery stage (M = 0.49, SD = 0.19) had a lower mean harvest index value than the clonal evaluation stage (M = 0.57, SD = 0.15), t (146) = -14.17, p = 0.000. The clonal evaluation trial showed significantly lower root dry matter content (M = 27.54, SD = 5.24) than the seedling nursery stage (M = 29.89, SD = 6.75); t (146) = 3.48, p = 0.001.
Using SNPs molecular markers for heterozygosity, the S1 generation showed a good proportion of heterozygotes at several loci across the genome, indicating that heterozygosity is very high in cassava, and several selfing cycles may be required to reduce it drastically. The heterozygosity indices of S1 progenies are shown in Figure 1. SNP analysis revealed percentage heterozygosity between 12% and 65% in the population. The average heterozygosity reported for the S1 families is shown in Table 7. The results showed that estimates of the heterozygosity index tended to increase and improve as a greater number of SNP markers were used. This suggests that an optimum number of SNPs should be determined to improve the precision of heterozygosity estimates in cassava. Figure 2 shows the distribution of S1 progenies based on their percentage heterozygosity estimated using SNP markers. The findings also demonstrated that a majority of the progenies, specifically over 55%, exhibited heterozygosity values below 25%. This suggests that there are quality materials to pass on to the next inbreeding generation, which will likely result in less heterozygosity in the subsequent breeding generation.
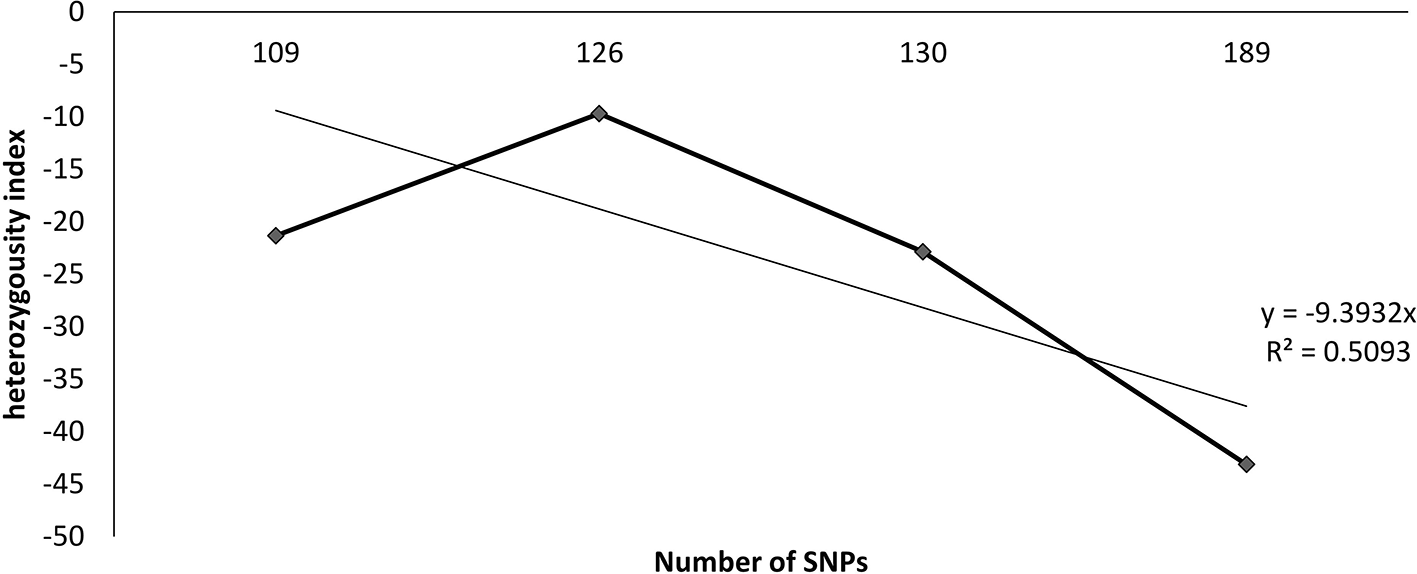
| Family | S0 | S1 | Heterozygosity index |
|---|---|---|---|
| TME 419 | 39.5 | 31 | -21.32 |
| TMS 01/1371 | 27 | 24.4 | -9.69 |
| TMS 30572 | 39.2 | 30.3 | -22.87 |
| TMS 98/0002 | 7.32 | 23.3 | 218.02 |
| TMS 98/0505 | 46 | 26.2 | -43.13 |
| Average | 31.8 | 27 | 24.21 |
Phenotypic measurements of key breeding traits were performed during the seedling and clonal evaluation trials. This study explored SNP markers to assess the heterozygosity of the top five elite clones that have been extensively used as parents in cassava breeding programs.
The inbreeding depression for fresh root yield, which is a very complex trait (involving many genes), was found to be higher than the values obtained for other traits evaluated in this study. These results suggest that a high ID was more likely for traits that were more quantitatively controlled (e.g., root yield) than traits controlled by fewer genes, for example, plant height. This agrees with the findings of Castro et al. (2006).
In the case of cassava mosaic disease, cassava bacterial blight, and cassava anthracnose disease, little depression was observed. This was probably because of additive genetic effects, in addition to the fact that pests and disease resistance are often affected by a few genes (including dominant or major gene effects). For example, previous molecular studies indicated that CMD resistance in two of the varieties, TMS98/0505 and TMS98/0002, used as parents, is mainly controlled by a single dominant gene resistance (Okogbenin et al., 2013).
These results suggest that breeders can explore inbreeding for these traits in the early stages of the selfing cycle within the breeding scheme, potentially enhancing their genetic progress. Some previous studies in cassava have reported disease resistance to CMD and CBB as being quantitatively controlled, with some being recessive. Furthermore, the low ID for pests and diseases shows that traits that are strongly influenced by additive genetic variance stand to benefit most, leading to genetic gain. Additionally, useful beneficial recessive traits can be effectively explored while simultaneously selecting undesirable recessive traits.
In general, the inbreeding depression values were higher in the seedling evaluation trial than in the clonal evaluation trial for fresh root yield, fresh foliage yield, harvest index, and dry matter content. The use of seeds versus stem cuttings may have influenced the results. In the seedling trial, cassava was raised from seeds, whereas stem cuttings were used as planting materials in the clonal trial. Seeds typically produce plants with fewer and smaller roots than those raised from stem cuttings (clonal stage) (FAO, 1977). Food reserves in stem cuttings aid in rapid establishment and improved performance. Therefore, it is important to explore later generations to evaluate inbreeding depression for these traits.
A paired samples t-test was performed to determine whether there was a statistically significant difference between seedling nursery and clonal evaluation stages for all traits evaluated. The results revealed a significant difference between the seedling nursery and clonal evaluation stages for all traits evaluated. The expression of the disease traits (cassava mosaic disease, cassava bacterial blight, and cassava anthracnose disease) was more severe at the seedling nursery stage than at the clonal evaluation stage. Likewise, morphological traits (vigor and plant height) performed better at the clonal evaluation stage than at the seedling stage. This could be because during the process of moving from the seedling evaluation stage to the clonal evaluation stage, genotypes with poor vigor were eliminated, while vigorous genotypes were selected for further testing. For yield traits, such as fresh foliage yield, fresh root yield, and harvest index, the use of stem cuttings may have provided better results in the clonal evaluation stage. The dry matter content was significantly lower at the clonal evaluation stage than at the seedling nursery stage. At the seedling nursery stage, the genotypes mainly had fibrous roots and a higher dry matter content than at the clonal evaluation, where tuberous roots and a lower dry matter content were observed.
Cassava has been demonstrated to be a highly heterozygous species (Kawano et al., 1978; Pujol et al., 2005). Inbreeding depression, such as that observed in this study, was therefore expected. ID is the result of a reduction in heterozygosity levels in loci with dominant gene effects and because of the increased frequency of expression of unfavorable alleles (Falconer, 1981; Miranda-Filho, 1999, Vencovsky and Barriga, 1992). Inbred lines are more suitable for breeding and genetic research due to their lack of dominance effect and reduced genetic load (undesirable alleles), making them superior parents.
Five S1 individuals, one genotype each from the families of TMS 98/0505, TME 419, and TMS 30572 and two genotypes from the family of TMS 01/1371 showing relatively low heterozygosity based on SNP data, were selected as parents for further selfing to assess ID. Therefore, these genotypes were further advanced to generate an S2 population for further study.
The impact of inbreeding on pests, diseases, and morphological and architectural traits in the S1 generation was non-severe. Notably, the presence of transgressive segregants in the population for key strategic traits of the crop is indicative of favorable allele combinations at the S1 stage. Therefore, the results demonstrate that inbreeding can be strategically explored in breeding for these traits to increase genetic gain and possibly identify recessive traits. At the early stages of the cassava breeding scheme, most key traits, including disease resistance, were not severely affected at S1. This presents an opportunity to identify major or a few genes associated with disease resistance and to fix them in early generations to generate valuable parent stocks. These parent stocks can then be used in crosses, enabling the exploration of both additive and non-additive variances for polygenic traits in later breeding cycles. Moreover, the strategic use of inbreeding allows for early selection of productivity traits once genetic stocks become available. This can significantly shorten the breeding cycle, especially when appropriate technologies are employed to rapidly multiply selected genotypes in larger yield trials. In summary, our findings suggest that inbreeding can be a valuable tool in cassava breeding for specific traits, offering potential benefits in accelerating genetic progress and identifying essential recessive traits. By incorporating inbreeding strategies and leveraging advanced technologies, cassava breeding programs can make significant strides in developing improved varieties that address the challenges of pests, diseases, and productivity in this vital crop.
The datasets presented in this study can be found in the figshare repository.
1. SNPs estimates for heterozygosity , DOIs: https://doi.org/10.6084/m9.figshare.26312278.v1 (Jiwuba, 2024b)
This project contains the following underlying data:
2. Inbreeding depression seedling and clonal evaluation trials.xlsx, Doi: https://doi.org/10.6084/m9.figshare.26317900 (Jiwuba, 2024a)
This project contains the following underlying data:
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Special thanks to the technical team working at NRCRI, Umudike, for their assistance during the field activities and data collection.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Agricultural researcher specializing in variety improvement, with expertise in breeding new crop varieties using both conventional and biotechnology approaches. Skilled in seed conservation, seed systems development, and enhancing genetic diversity for improved agricultural productivity and sustainability.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Application of biotechnological tools for biotic and abiotic stress resistance in cassava and other root crops
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 27 Aug 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)