Keywords
soybean, yellow mosaic virus tolerance, simple sequence repeat marker, genetic resistance, Rsv1-h gene
This article is included in the Plant Science gateway.
Twenty soybean genotypes were screened for Soybean yellow mosaic virus (YMV) identification which were collected from the Genetics and plant breeding farm laboratory.
To evaluate genetic studies of the chosen materials, the use of a net house assay upon sap inoculation made it easier to determine the trend of the Area Under Disease Progress Curve (AUDPC) in a controlled environment. Younger leaves from these plants were gathered and used for molecular analysis.
Six genotypes were identified with semi-resistant disease reactions. Yellow mosaic virus-resistant and susceptible soybean genotypes were distinguished using a soybean “Rsv1-h” gene-based primer pair. To discover the existence of the Rsv-1-h gene responsible for YMV resistance, a genetic diversity panel of 15 soybean genotypes was investigated with two simple sequence repeat (SSR) markers. The BARCSOYSSR_13_1115 and BARCSOYSSR_13_1173 primer sets were employed to validate the desired genes in these genotypes.
The genotypes HIHS-WIHS, MINA-HAI, Shohag, G-2120, BRAGG, and BS-3 presented the major gene Rsv1-h, which is said to be essential for yellow mosaic virus resistance and can provide better yield also than other genotypes. The six soybean genotypes with the Rsv1-h gene may be resistant to mosaic virus and could be exploited as a source of improved breeding material in the future.
soybean, yellow mosaic virus tolerance, simple sequence repeat marker, genetic resistance, Rsv1-h gene
Soybean (Glycine max) is one of the significant legume crops in today’s world, whose protein content is higher than other legumes.1,2 29.7% of the total vegetable oil is produced from soybeans and its major producers are the USA, Brazil, Argentina, and China.3 As it includes 36.6 g of protein, 19.9 g of total fat, 30.2 g of carbohydrates, 9.3 g of dietary fibre, and 15.7 mg of iron per 100 g of seeds, it is nutritious for human and animal consumption.4 Soybean is cultivated in 62,870 hectares (ha) of the total cultivable area in Bangladesh, from which we get nearly 96,921 tons of soybean per year.5 However, many biotic and abiotic factors adversely affect the growth and productivity of soybeans. Among them, the soybean business faces a severe threat from the soybean yellow mosaic virus (YMV), one of the most notorious agents of yield loss among 100 viruses.6 The disease can spread from leaves to seeds during severe infestation which deteriorates the quality of the seeds.7 The economic loss caused by this disease is 30-50%; however, it may go up to as high as 80% in extreme cases.8 As YMV is a viral disease, its control through chemical or cultural practices is neither effective nor environmentally friendly. So, implementation of genetic resistance can be an ideal option for soybean yellow mosaic virus management. For such an approach to be effective, it is crucial to understand the genetic control of the disease.9 The challenge in identifying resistant soybean plants is frequently hampering the progress in the breeding of soybeans in our country. If suitable screening methods are not used, selection may result in picking plants with pseudo-resistance, as YMV is delivered by whiteflies and their distribution in the field may not be uniform.10
The generation of YMV-resistant genotypes is substantially facilitated by selection using indirect methods, such as gene linked-molecular marker(s), which may help to overcome the limitations of field screening. Microsatellites are among the most widely used molecular markers because of their high degree of polymorphism, co-dominance nature, chromosome specificity, and dependability.11 Simple Sequence Repeat markers, commonly known as SSR markers, have been utilized to locate YMV resistance gene-linked markers in soybean.12 Field screening and subsequent selection can be made easier by using these marker(s) for YMV resistance. Marker-assisted selection (MAS) is a common strategy in molecular breeding efforts for identifying YMV in soybeans.13 The species of soybeans YMV prevalent in Northern Bangladesh differed from those prevalent in Southern Bangladesh.14 For this reason, the resistance of genotypes may also vary from one region to another region depending upon the strain of the virus prevalent in our country. For the adoption of successful methods for soybean YMV resistance breeding, it is necessary to investigate and unequivocally establish the genetic control of the disease. For this reason, the present experiment was done to evaluate several soybean germplasms to identify YMV disease resistance.
The field experiment was conducted at the experimental farm of the Department of Genetics and Plant Breeding, Bangladesh Agricultural University (BAU). It was arranged in a randomized complete block design with three replications. For molecular data analysis, soybean plants were grown in a controlled environment in the Genetics and Plant Breeding net house. The laboratory work has been carried out at the Plant Breeding Division of Bangladesh Institute of Nuclear Agriculture (BINA), Mymensingh, Bangladesh. The whole experiment was done in July 2019-June 2020.
Nineteen exotic germplasm and one BAU variety were collected for this experiment. In a controlled environment, younger leaves at 3 to 4 stages of soybean were rubbed with inoculation sap (viral solution) for 3 to 4 times at 3 days intervals. The inoculation sap was prepared by homogenizing young leaves with typical mosaic symptoms of virus-infected plants. Leaf inoculation was done to infect younger plants. Individual plants were scored three times for the appearance and type of symptoms at 7-, 10- and 14-days post-inoculation (dpi) with day 0 being the first inoculation date.
Data were recorded for morphological studies and genetic analyses of the selected soybean genotypes. Plants with no symptoms, partial symptoms and systemic mosaic symptoms were rated as 0, 1 and 2 according to the severity of the disease.15 The scoring of leaves is shown in Table 1.
Disease severity data were used to compute the area under disease progress curve (AUDPC) described by Campbell and Madden (1990).
Where, n = number of successive readings,
yi = disease severity at time i,
ti = number of days after the observation on assessment date i.
AUDPC was drawn for each plant in each replicate using the mean plot disease score at each rating date.16 No vector was allowed for any kind of transmission from plant to plant. Mosquito nets were used to cover up the inoculated leaves before taking data for AUDPC.
Primer used for amplification of soybean yellow mosaic virus
In the net house experiment, leaf inoculation was allowed to infect younger plants as described by Chen et al., 2015.17 Infected leaves of earlier growth stages were collected from those soybean cultivars and were used as plant materials for molecular analysis. Two sets of primer were selected on the basis of the intensity of bands, presence of smearing, consistency with individuals and potential for population discrimination.
Isolated genomic DNA was tested for quality and quantity following standard procedure. The polymerase chain reaction (PCR) amplification profile consisted of initial denaturation at 94°C for 2 minutes followed by 35 cycles consisting of denaturation, primer annealing and extension at 94°C, 60°C and 72°C, respectively, for 30 seconds to 1 minute each. PCR cocktail made with 5 μL master mixture, 1 μL of template DNA, 1 μL of each forward and reverse primer and 2 μL of Nuclease-free water, which was amplified in a standard thermocycler.
Analysis of SSR data
Gene diversity was calculated using Roldan-Ruiz et al. (2000)’s formula:
where, GDi is the gene diversity of marker ‘i’, fi is the frequency of the amplified allele (band presence), and 1 - fi is the frequency of the null allele. The GD for a locus can range from 0 to 0.5 using this calculation.The presence and absence of DNA band were considered for identifying the presence or absence of the gene of interest. Polymorphism information content (PIC) values described for self-pollinated species were calculated as follows:
Where, Pij is the frequency of the jth allele for ith marker and summed over n alleles.
We did not obtain high-quality amplification from every soybean genotype. Among all polymorphic markers, polymorphism information content (PIC) was highest (0.61) in BARCSOYSSR_13_1173 and lowest (0.11) in BARCSOYSSR_13_1115 (Table 2). The list of the SSR markers and their PIC values are as follows:
• BARCSOYSSR_13_1115 whose PIC value is 0.11 and major allele frequency is 0.8113
• BARCSOYSSR_13_1173 whose PIC value is 0.61 and major allele frequency is 0.625
Both of these markers’ product size were 299 bp.
Performance of soybean genotypes after virus inoculation
Asymptomatic plants were scored as 0; whereas plants with partial symptoms and plants with mosaic symptoms (50-80%) were scored as 1 and 2 accordingly (Figure 1).

The percent infection and AUDPC for each genotype were calculated from the scores obtained after virus inoculation at 7, 10 and 14 dpi (days post inoculation) and shown in Table 3. The high value of % infection and AUDPC indicated susceptibility, and the low value stated resistance. For this reason, MINA-HAI, Shohag, HIHS-WIHS, BS-3, BRAGG might be considered as highly resistant; Davis, Lokon, G-10180, Asset 93-19-5, G-2120 as moderately susceptible; AGS-66, TAINANS as highly susceptible (Table 3).
Five genotypes were found, which showed high resistance of all. The relationship of Area under Disease progress curve (AUDPC) and %Infection in soybean genotypes is shown in Figure 2.
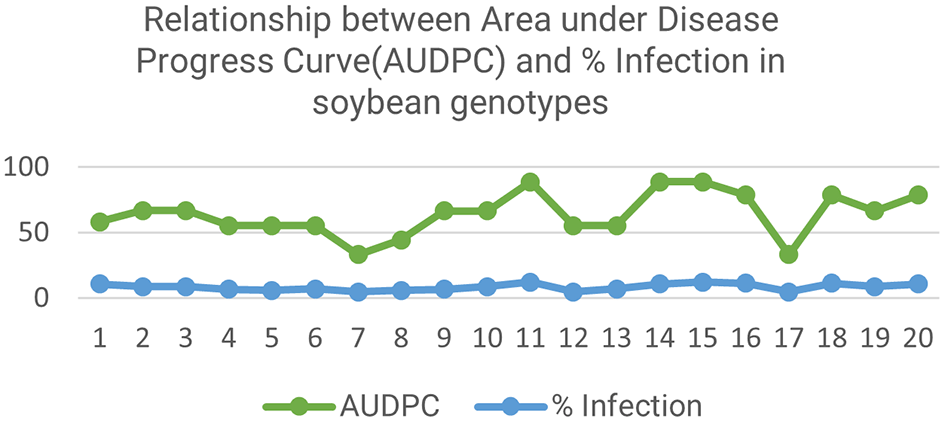
Here, the highest percent infection (88.66%) was observed for BS-13, AGS-278, and Jayawiyaja and lowest for MINA-HAI and Asset-93-19-1 (33.33%). BS-13 and Jayawiyaja showed the highest AUDPC value (12.1) and MINA-HAI, HIHS-WIHS, and Asset-93-19-1 showed the lowest (4.8).
Molecular detection using specific primer
Though the disappearance and reappearance of viral symptoms often mislead us, pathological identification via AUDPC helped in the virus detection system. We emphasized Rsv1 gene identification, which has a significant role in resistant regulation in soybean. Two sets of primers were used to detect the yellow mosaic virus gene in soybean genotypes which is responsible for yellow mosaic virus resistance. The highly susceptible genotypes were cut down and SSR markers were used to detect desired genes in the 15 soybean genotypes. Individual soybean plants were screened for two simple sequence repeat (SSR) markers named BARCSOYSSR_13_1173 and BARCSOYSSR_13_1115 that flank the Rsv1 locus.
From Figure 3, it was observed that while using the BARCSOYSSR_13_1173 SSR marker, six genotypes namely Mina-hai, Shohag (Pb-1), G-2120, HIHS-WIHS, BS-3 and BRAGG showed fragments at the expected size (299 bp) based on the simple PCR detection system. The viral resistance system is a complex mechanism that provides resistance in a semi-persistent manner, so a certain degree of resistance can be expected if the significant gene is present in a genotype. A remarkable variation was noticed in 15 genotypes (Figure 3). The PCR-based results indicated that a fragment of 300 bp was absent in most of the genotypes as the primers were gene-specific. Mina-hai, Shohag, G-2120, HIHS-WIHS, BS-3, BRAGG carried Rsv1-h gene against Soybean yellow mosaic virus (Table 3). The rest of the genotypes showed the absence of a 299-300 bp fragment, indicating that these genotypes were susceptible to SYMV. Besides this, it was observed that none of the genotypes showed fragments at the expected size (299bp) using the BARCSOYSSR_13_1115 primer.
Viral symptom scoring was performed at 7, 10 and 14 days after inoculation of the virus. MINA-HAI, Shohag, HIHS-WIHS, BS-3, BRAGG showed resistance as their infection rate and AUDPC value were low. On the other hand, AGS-66 and TAINANS were highly susceptible because of their high value of % infection rate and AUDPC value. Davis, Lokon, G-10180, Asset 93-19-5, G-2120 was partially susceptible due to their medium level of infection rate and AUDPC value. The phenotypic performance of soybean genotypes regarding yield and yield-contributing traits partially supported this hypothesis. Stewart et al., (2013) inoculated virus to a number of genotypes and calculated % infection and AUDPC at 7, 10 and 14 dpi. They found that the resistant genotype showed 0% infection and the susceptible one showed 100% infection.15 Molecular work was also performed to ensure the scoring results. The identified resistant genotypes were expected to carry the rsv1-h gene.
For the purpose of screening individual soybean plants, we looked for two simple sequence repeat (SSR) markers that border the Rsv1 locus: BARCSOYSSR_13_1173 and BARCSOYSSR_13_1115. This marker was also utilised by Lee et al. (2013) to identify yellow mosaic virus resistance in soybean.18 The Rsv1 - h gene in cultivar Suweon 97, which confers resistance to SMVs, was found to be mapped to a 97.5-kb location (29,815,195–29,912,667 bp on chromosome 13) in the Rsv1 locus by Ma et al. (2016). This observation gave rise to further questions regarding the molecular basis of variations in resistance alleles in this specific locus.19 The dominant Rsv1 gene has been assigned to chromosome 13 (molecular linkage group F). While Rsv1 has been the subject of extensive research and discussion among scientists worldwide, as evidenced by the numerous citations, it is important to acknowledge the possibility of the existence of distinct viral strains in Bangladesh that require systematic attention in our local environment.
The six genotypes viz. Mina-hai, Shohag, G-2120, HIHS-WIHS, BS-3, BRAGG having the expected fragment of 299-303 bp using BARCSOYSSR_13_1173 primer sets which were responsible for resistance regulation against soybean yellow mosaic virus. The Rsv1 - h gene enables soybean plants to maintain better stress conditions. Zheng et al. (2014) identified two genomic-simple sequence repeat (SSR) markers, BARCSOYSSR_13_1114 and BARCSOYSSR_13_1136, which are located on either side of the RSC3Q and are associated with the Rsv1 area.20 A quantitative real-time PCR analysis of the candidate genes identified five genes that were possibly implicated in soybean YMV resistance: Glyma13g25730, 25750, 25950, 25970, and 26000. The cloning of the RSC3Q resistance candidate gene and the utilization of marker-assisted selection (MAS) in YMV resistance breeding could both benefit from these findings.
Further extensive molecular and serological studies may be helpful in understanding the Rsv1-h gene-mediated resistance regulation of soybean yellow mosaic virus.
Eventually, six genotypes have been found to be semi-resistant out of twenty genotypes. The genotypes HIHS-WIHS, MINA-HAI, Shohag, G-2120, BRAGG and BS-3 all have the major gene Rsv1-h, which is considered necessary for yellow mosaic virus resistance. They also have produced higher yields than other genotypes. These six soybean genotypes could be used in the future as a source of improved breeding material. During virus indexing at the field level, five genotypes showed highly susceptible expression of the YMV disease. An effort has been made in this research to identify genotypes that are resistant to YMV by using disease indexing and SSR markers. These genotypes could be used as parents in future crossing programs to develop YMV-resistant soybean cultivars through molecular breeding. The improved yellow mosaic virus-resistant genotypes could be used as a barrier against disease transmission to additional locations, perhaps increasing soybean yield in our country.
Figshare: Underlying and extended data for ‘Detection of yellow mosaic virus resistance in Soybean (Glycine max L.) genotypes for yield and related traits’, https://www.doi.org/10.6084/m9.figshare.25918495.v3. 21
This project contains the following underlying and extended data:
• Scoring of soybean leaves
• List of SSR Markers used for Soybean yellow mosaic virus identification
• Performance of soybean genotypes after virus inoculation
• Figure 01. Symptoms of yellow mosaic virus plants <.jpg format>
• Figure 02. Relationship between Area Under Disease Progress Curve and % Infection in soybean genotypes <.jpg format>
• Figure 3. The banding pattern of 15 soybean genotypes using the BARCSOYSSR_13_1173 SSR marker <.jpg format>
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
We acknowledge the support of the Bangladesh Institute of Nuclear Agriculture for the molecular analysis of the soybean leaf DNA samples.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Plant Pathology, Plant Virology, Plant-microbe interaction, Molecular Plant Pathology, Virus Diagnostics.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Plant Virology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Host Plant Resistance to Pathogens, Biological Control of plant diseases
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
|
Version 1 30 Aug 24 |
read | read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)