Keywords
SARS-CoV-2, Variants of Concern, Genomic Epidemiology, Vaccination, Children, Pakistan, Phylogenetics, Subclinical Transmission
This article is included in the Genomics and Genetics gateway.
This article is included in the Emerging Diseases and Outbreaks gateway.
This article is included in the Coronavirus (COVID-19) collection.
Pakistan experienced six COVID-19 waves with varying SARS-CoV-2 variants. Vaccination campaigns began in 2021, in an age-stratified manner starting with the older-aged individuals, yet the relationship between vaccination status and variant transmission remains unclear. We investigated SARS-CoV-2 phylogenetic diversity in relation to age and vaccination status, with a focus on children.
We analyzed 569 SARS-CoV-2 genome sequences collected across Pakistan from May 2021 to October 2022. Metadata including age, sex, and vaccination status were retrieved from GISAID. Additional data such as viral load was available for a set of 143 pediatric samples (< 18 years) sequenced at Aga Khan University Hospital (AKUH), Karachi, and was analyzed separately to gain further insights. Phylogenetic analyses were conducted using the Augur pipeline and visualized via Auspice. Viral loads were inferred from Ct values of SARS-CoV-2 PCR diagnostic assays.
Among the 569 genomes, Delta (45.2%) dominated in 2021 and Omicron (45.9%) in 2022. Of these, 71% were from individuals <18 years; 29.3% were vaccinated. Variant distribution showed no significant difference between vaccinated and unvaccinated individuals (p=0.75), nor between age groups (<18 vs ≥18 years; p=0.60). Pediatric samples from AKUH Karachi mirrored national trends, with Delta predominating in 2021 and Omicron in 2022. Omicron variants were associated with higher viral loads (p<0.001).
SARS-CoV-2 variant distribution and genomic divergence were comparable across vaccination status and age groups. In a population with high subclinical transmission and delayed pediatric vaccination, our findings highlight the stability of variant spread irrespective of host immunity status, suggesting the role of widespread prior exposure in shaping transmission dynamics.
SARS-CoV-2, Variants of Concern, Genomic Epidemiology, Vaccination, Children, Pakistan, Phylogenetics, Subclinical Transmission
The COVID-19 pandemic, caused by SARS-CoV-2, led to unprecedented global health, social, and economic disruptions between 2020 and 2022, with ongoing endemic transmission into 2025. In Pakistan, approximately 1.6 million cases and 30,700 deaths were reported to the World Health Organization (WHO) from March 2020 to April 2023 (WHO, 2024c).
Since its emergence, SARS-CoV-2 has evolved into a range of genetically distinct variants. Those posing increased risks to global public health were categorized by the WHO as Variants Under Monitoring (VUM), Variants of Interest (VOI), and Variants of Concern (VOC) (WHO, 2023a, 2023b). VOCs are defined by their enhanced transmissibility, potential to cause more severe disease, escape immune responses, or reduce the efficacy of public health interventions, including vaccines (WHO, 2023b). From May 2021, the WHO adopted Greek letters to label key variants, including Alpha, Beta, Delta, and Omicron—several of which have been associated with vaccine breakthrough infections (Jamal et al., 2022; WHO, 2021).
SARS-CoV-2 genomic divergence is driven by mutation, recombination, and genetic drift (Dvořák, Jahodářová, Stanojković, Skoupý, & Casamatta, 2023). Global sequencing efforts have enabled high-resolution tracking of variant emergence and spread, informing public health strategies. Emerging evidence suggests that SARS-CoV-2 genomes began stabilizing after widespread vaccine rollout, as shown by reduced substitution rates and viral entropy (Ashraf et al., 2023).
Vaccination against SARS-CoV-2 has been pivotal in mitigating severe disease and mortality. During the first year of vaccine deployment (December 2020–December 2021), COVID-19 vaccines were estimated to have prevented 19.8 million deaths globally (Watson et al., 2022). In the WHO European Region alone, 1.4 million lives were saved between December 2020 and March 2023 (WHO, 2024b). In Pakistan, vaccines were introduced in 2021, initially prioritizing healthcare workers and the elderly. The national program deployed a mix of mRNA, inactivated virus, and viral vector vaccines. Reported vaccine effectiveness (VE) ranged from 33.8% to 67.4% depending on the platform and circulating variant (Camacho-Moll et al., 2024; Firouzabadi, Ghasemiyeh, Moradishooli, & Mohammadi-Samani, 2023) (Nisar et al., 2023). During the Delta wave, VE against infection was lower for inactivated vaccines (Khan et al., 2023), though protection against severe disease remained robust across platforms (WHO, 2024a). While vaccines clearly reduced hospitalizations and deaths (Wang et al., 2023) (Mwendwa et al., 2024), it remains unclear whether vaccination status altered the spectrum of infecting SARS-CoV-2 variants.
Additionally, there is limited data on SARS-CoV-2 variant dynamics in children in Pakistan. A multicenter study conducted between March 2020 and December 2021 reported an 18.6% mortality rate among hospitalized pediatric COVID-19 patients (Abbas et al., 2023). This study noted the highest pediatric case surge and mortality early on during the Delta wave (July–September 2021), but did not explore variant-specific susceptibility in children.
In this context, we investigated SARS-CoV-2 genomic diversity in Pakistan during May 2021–October 2022, spanning the Delta and Omicron waves across the different provinces of Pakistan. We examined circulating variants in vaccinated and unvaccinated individuals, with particular attention to children under 18 years. By analyzing national sequence data alongside pediatric cases from Karachi, we aimed to assess whether age or vaccination status influenced variant distribution or viral divergence.
This was a retrospective genomic epidemiological study conducted using SARS-CoV-2 genome sequences from Pakistan, spanning May 2021 to October 2022.
This study was conducted in accordance with the ethical standards set forth in the Declaration of Helsinki. Ethical approval was obtained from the Ethics Review Committee of Aga Khan University, which granted a waiver of written and verbal informed consent for this retrospective analysis (Approval No. 2021-6316-18156 and 2022-6871-22433). The waiver was justified because this was a retrospective analysis using secondary data, where re-contacting participants to obtain consent was neither feasible nor practical. Such an approach is consistent with ethical guidance for studies involving anonymized or previously collected data, where the risks to participants are minimal. All data was de-identified prior to analysis to ensure confidentiality.
We retrieved all available SARS-CoV-2 genome sequences (FASTA format) and associated metadata for Pakistan from the GISAID database (https://www.gisaid.org). Filters were applied to include only sequences with “high coverage” and “complete collection date.” From this dataset, 569 sequences were selected for downstream analysis based on the availability of vaccination status in the metadata.
Additionally, data for SARS-CoV-2–positive samples from individuals <18 years of age tested at Aga Khan University Hospital (AKUH), Karachi, were included for pediatric-focused analysis (n = 143). The specimens were selected as previously described by Nasir et al. (Nasir et al., 2023), by identifying the first 10 positive samples per day. Inclusion criteria included a PCR cycle threshold (Ct) value ≤35 using the Cobas SARS-CoV-2 assay. Samples with Ct >35 or duplicates from the same individual were excluded.
Vaccination status was assigned based on metadata and age. Individuals ≥18 years were considered vaccinated or unvaccinated based on reported status. Since COVID-19 vaccines were not available to individuals <18 years in Pakistan until late 2021 (GoP, 2021), all pediatric cases (<18 years) during the study period were considered unvaccinated unless otherwise specified.
Phylogenetic analysis was conducted using the Nextstrain Augur pipeline and visualized via the Auspice platform (https://auspice.us). Sequences were aligned to the SARS-CoV-2 reference genome (NC_045512.2) using MAFFT. A maximum-likelihood phylogenetic tree was constructed using IQ-TREE2, employing the General Time Reversible (GTR) substitution model. Ancestral sequence inference and molecular dating were performed within Augur, and Nextstrain clade assignments were included. Divergence values were extracted from the JSON output of Augur for downstream statistical comparisons. Visualization filters (e.g., by age group, vaccination status, and time) were applied in Auspice for subgroup analysis.
Statistical analyses were performed using IBM SPSS v24 and GraphPad Prism v8.0.1. The Kolmogorov–Smirnov test was used to assess normality of divergence data. Chi-square, unpaired samples t-test and Mann Whitney U tests were used to analyze for significant differences. For comparisons involving more than two groups (e.g., Ct values across variants), the Kruskal–Wallis test followed by post-hoc pairwise comparisons was used. A p-value < 0.05 was considered statistically significant.
We wanted to understand the effect of COVID-19 vaccination on the transmission of SARS-CoV-2 variants in Pakistan by examining the genomic epidemiology of strains circulating in 2021 and 2022. The 569 SARS-CoV-2 genomes we analyzed were those available in GISAID for samples collected between May 2021 and October 2022 and which had metadata available including COVID-19 vaccination status (Supplementary table 1). Overall, Omicron and Delta variants were dominant at 45.87% and 45.17% respectively ( Table 1). 54.1% of the strains had been collected in 2021 of which the dominant variant was Delta (82.8%). The remaining (45.9%) strains were from 2022 when Omicron was dominant (92%). 47.1% genomes were collected from females. The greatest number of sequences (71%) were from strains collected from children (<18 years, p < 0.001).
Of the COVID-19 cases studied for vaccination status, there were 29.3% (n = 167) from vaccinated individuals and 70.7% (n = 402) from unvaccinated individuals ( Table 2). Most (69.5%) of the vaccinated persons were in the 18-50 years old category (p < 0.001). Most of genomes studied from unvaccinated COVID-19 cases were from children aged <18 years.
| Age category (years) | Total (n = 566) n(%) | Unvaccinated n(%) | Vaccinated n(%) | P-value |
|---|---|---|---|---|
| <18 | 402 (71.0%) | 399 (70.5%) | 3 (0.5%) | <0.001 |
| 18-50 | 116 (20.5%) | 2 (0.4%) | 114 (20.1%) | |
| >50 | 48 (8.5%) | 1 (0.2%) | 47 (8.5%) |
The SARS-CoV-2 genome submission were from across Pakistan. Of the 569 samples, most (38.7%) were from Islamabad (Islamabad Capital Territory, ICT), followed by Sindh (32.7%), Punjab (10.5%), Kyber Pakhtunkhwa (8.8%), Baluchistan (2.3%) and the rest from Gilgit-Baltistan and Azad Jammu and Kashmir ( Figure 1).
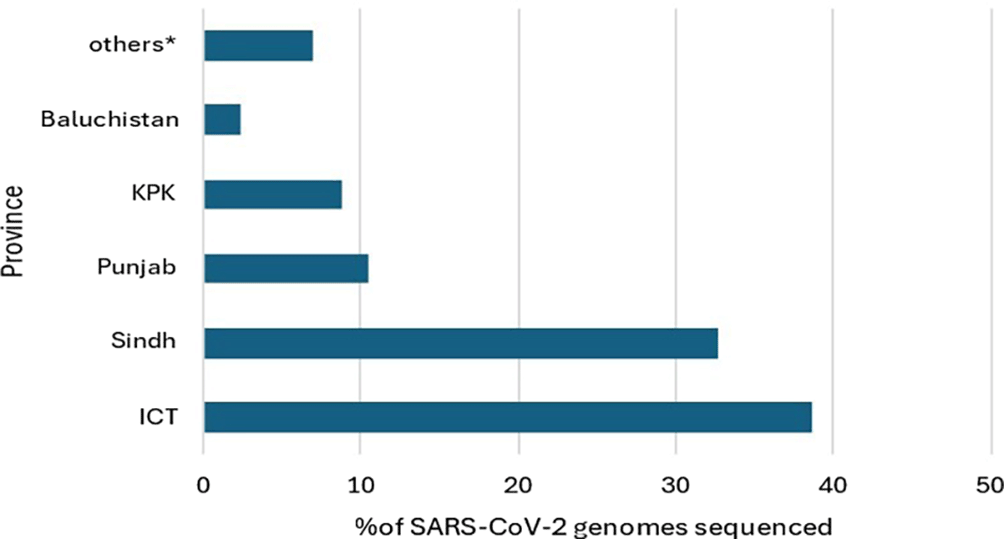
Graph shows geographical distribution of SARS-CoV-2 genomes downloaded from GISAID for phylogenetic analysis. Samples were from the provinces of Pakistan; ICT – Islamabad Capital Territory, KPK - Khyber Pakhtunkhwa. *includes samples from Gilgit-Baltistan and Azad Jammu Kashmir provinces as well as those for which location was unavailable.
The phylogenetic diversity of SARS-CoV-2 genomes sequences during the period of study showed the divergence of genomes between the Delta variants in 2021 and Omicron variants in 2022 was evident ( Figure 2A). The most common Delta lineage was B.1.167.2 (53.7%; n = 137) followed by AY.108 (24%; n = 60 (Supplementary Table 2)). In 2021 there were 1.3% Wildtype, 3.9% Alpha and 3.2 % Beta strains. In 2022, within the Omicron lineage BA.5.2 was prevalent (34.5%, n = 87). Importantly, isolates from both unvaccinated and vaccinated groups were found amongst the SARS-CoV-2 lineages identified. We further compared the occurrence of variants between vaccinated and unvaccinated groups. Strain divergence is visualized through a scatterplot and a similar pattern was observed in both vaccinated and unvaccinated cohorts ( Figure 2B). Genetic differences between vaccinated and unvaccinated individuals did not show any statistically significant difference (p = 0.75, unpaired samples t-test), (Supplementary Table 3).
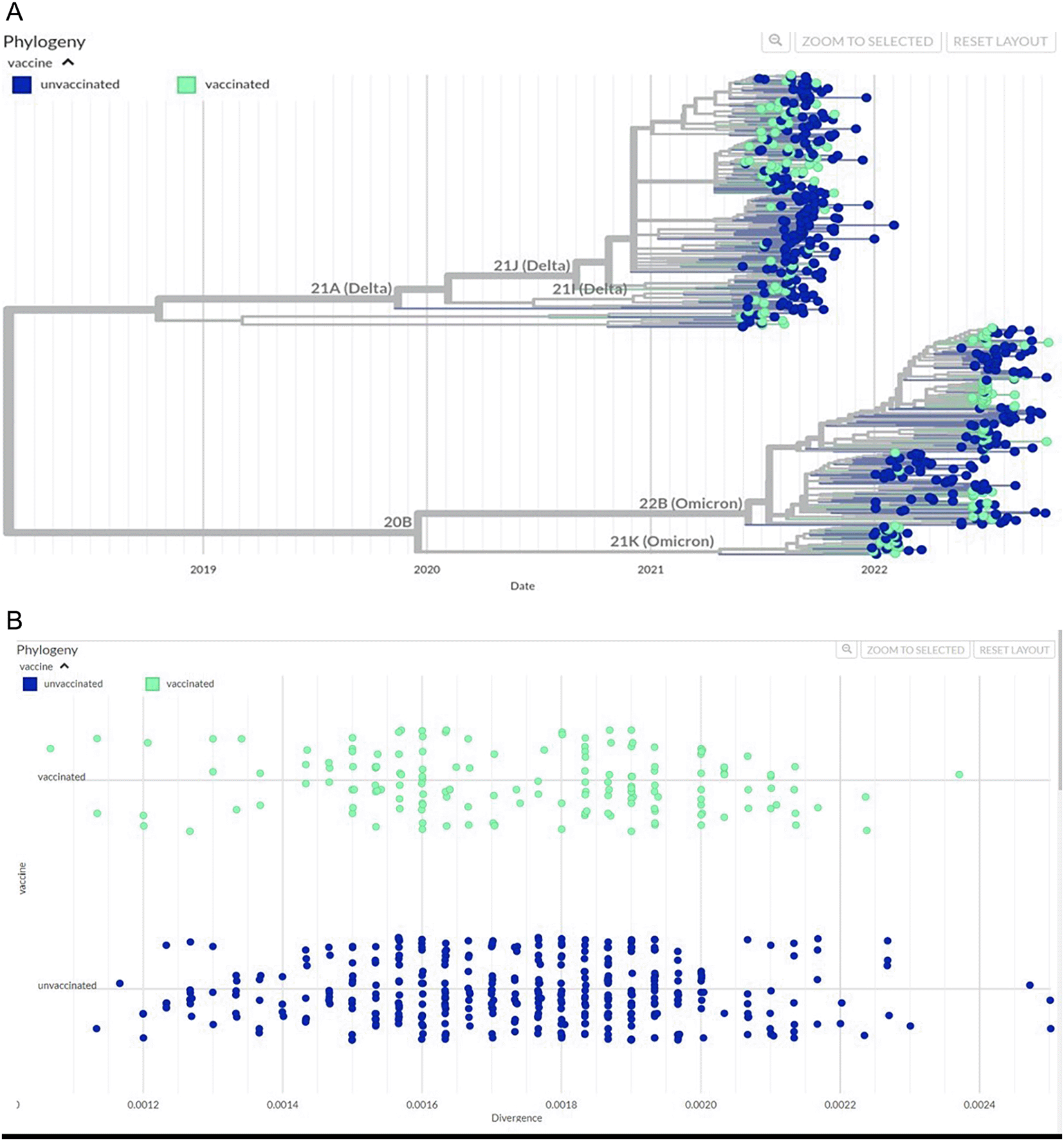
569 SARS-CoV-2 genomes were analyzed using the Augur pipeline. (A) The graph shows the diversity of SARS-CoV-2 genomes collected from unvaccinated (dark blue dots) or unvaccinated persons (light green dots) respectively. The X-axis shows samples along the timeline of the pandemic from 2019 to 2022, while the Y axis shows the two main branches of the phylogenetic tree. (B) The scatterplot shows genetic divergence between SARS-CoV-2 strains from either unvaccinated (dark blue) or vaccinated (light green) individuals. The X-axis of the graph shows divergence values of each sample in the cohort, and Y-axis shows the groups. The data was downloaded from GISAID (gisaid.org).
Next we compared the occurrence of variants across different age groups. Delta and Omicron were the most abundant variants across all age groups ( Figure 3A) and age-wise comparison of the strains showed a distribution of the prevalent SARS-CoV-2 variants (Beta, Alpha, Delta and Omicron) across older and younger age groups, as divided between those below 18 years and those aged 18 years and above ( Figure 3B, supplementary table 3). We divided the dataset for cases less than 18 years and more than 18 years. The statistical test between the two age categories did not give a significant difference in terms of divergence. Genomic divergence analysis of variants with age also displayed the spread of those <18 years and >18 years old cases to be similar ( Figure 3B). Also, unpaired samples T-test on the genomic divergence values did not give a significant statistical difference between the two groups (p-value = 0.6038, supplementary table 3).
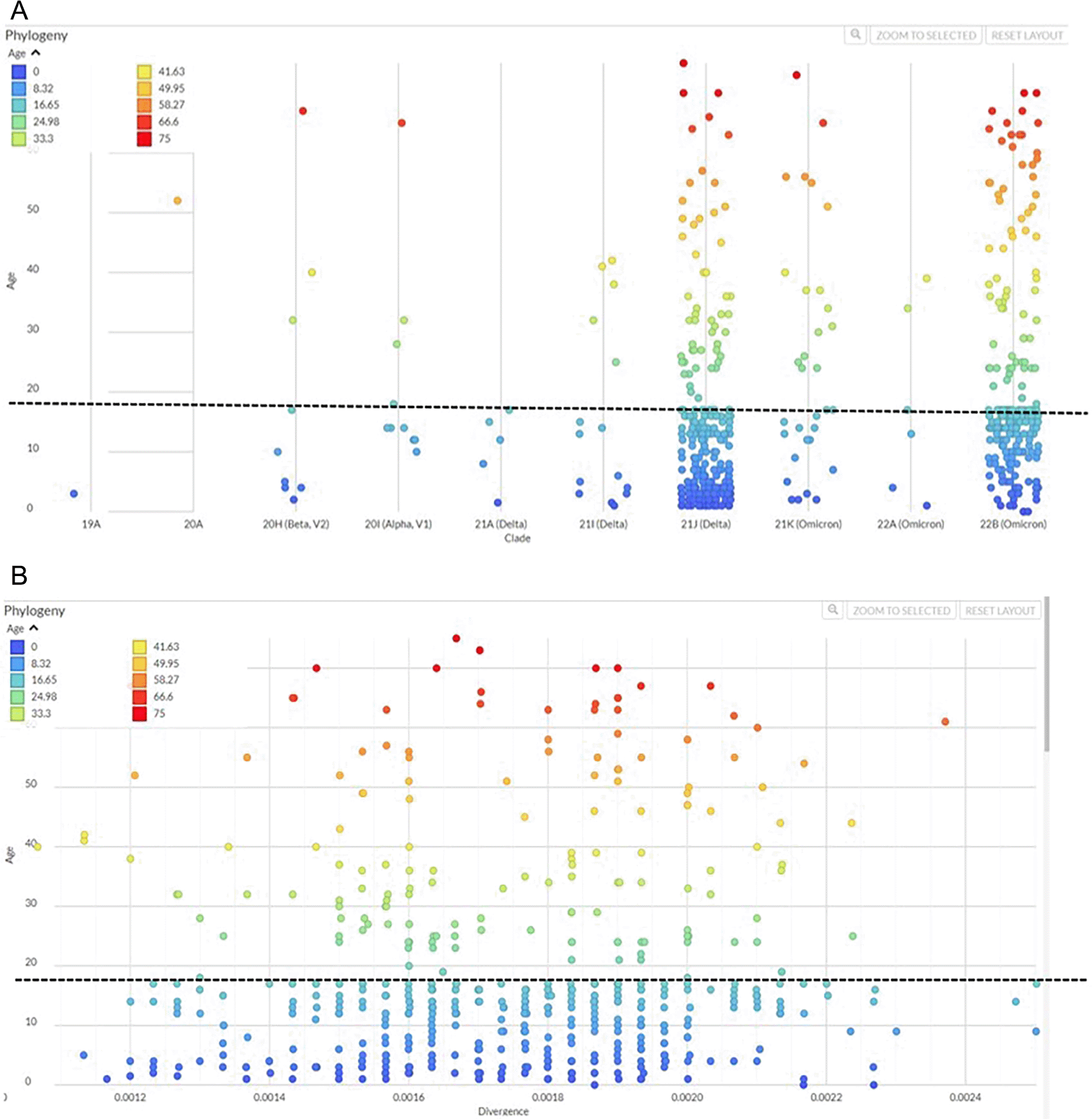
We compared the occurrence of SARS-CoV-2 variants based on the age of cases. (A) The graph depicts Next clade-based phylogeny of strains from individuals according to their age. The scatter plot shows the clade on the X-axis and age (years) on the Y-axis as represented by the dots for each individual.
(B) Strains were plotted to show genetic divergence as per age of individuals. SARS-CoV-2 divergence between samples is shown on the X axis whilst the Y-axis shows age. The dotted horizontal line marks at age 18 years to give a visual comparison of individuals aged less than 18 years and those above 18 years.
There has been limited information on COVID-19 in children aged <18 years and we now focused on this age group by comparing trends based on genomic sequences available countrywide and compared with those from our facility-based testing in Karachi. Of the SARS-CoV-2 genomes analyzed from across Pakistan, 402 were from those aged <18 years with 218 isolated in 2021 and 184 from 2022. In 2021, 89.9% of strains from this younger age group comprised Delta variants, and in 2022, Omicron was the most prevalent (90.2%) ( Figure 4A). We then focused on SARS-CoV-2 isolates received at the Aga Khan University Hospital as more information including, viral loads (Ct values of diagnostic PCR testing) were available. Of the 143 isolates collected from the under 18 year-olds with COVID-19 at our institute, 103 were from 2021 and 40 from 2022. Of these, 70% were from outpatient cases and 30% were from inpatients COVID-19 patients. We observed the dominant variants to be Delta in 2021 (57.3%) and Omicron in 2022 (100%) ( Figure 4B). Unfortunately, data on clinical disease severity of all individuals was not available.
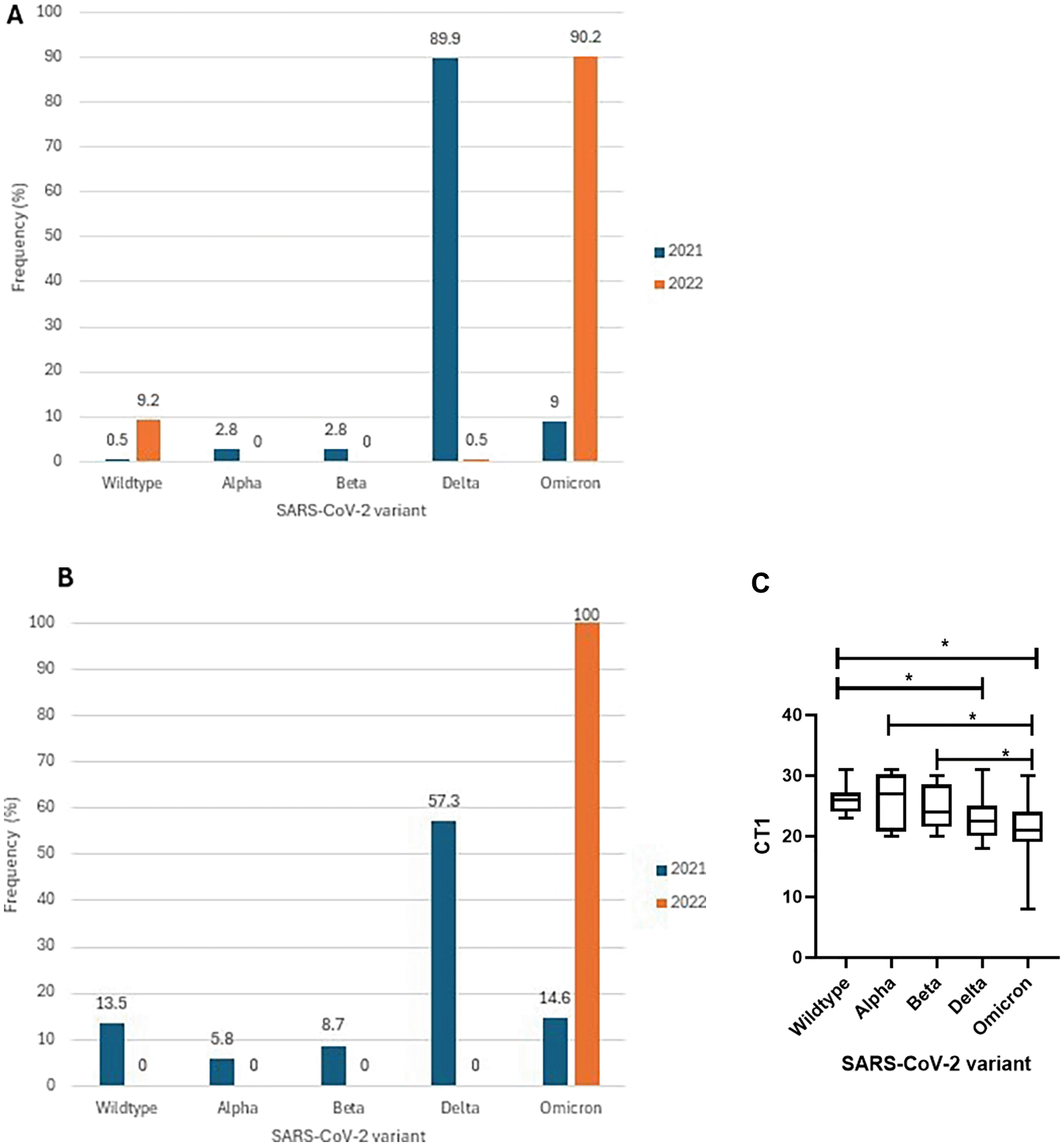
We first identified Wildtype and variants Alpha, Beta, Delta and Omicron amongst the strains studied. (A) The graph shows the frequency of variants in 402 genomes sequenced from children (<18 years) within the study cohort. The graph depicts the frequency of isolates from 2021 (n = 218) and 2022 (n = 184). (B) Identification of VOC from children diagnosed at AKUH. The graph depicts the frequency of 143 variants identified (2021 n = 103; 2022 n = 40). (C) The SARS-CoV-2 diagnostic PCR cycle threshold value was compared between the variants identified in those aged under 18 years at AKUH. Kruskal-Wallis test shows the medians vary significantly among the 5 groups, P = 0.0002. Mann-Whitney U test showed significant differences between Wildtype and Delta p=0.0007; Wildtype and Omicron p = 0.0001; Alpha and Omicron p-0.0449; Beta and Omicron p-0.0249; *CT1 denotes cycle threshold of orf1ab gene target. *p <= 0.05 was considered significant.
Further, we examined the SARS-CoV-2 viral loads of 143 AKUH specimens from those aged <18 years and found SARS-CoV-2 viral loads (as assessed by Ct values), to differ significantly between variants (Kruskal Wallis p value = 0.0002). There was a higher Ct in wild type compared with VOC Delta (p = 0.0007) and Omicron (p = 0.0001) ( Figure 4C). This indicated higher viral loads in cases involving VOCs as compared with Wildtype strains. There was no significant difference between Wildtype and Alpha, p = 0.6653; Wildtype and Beta, p = 0.3239; Alpha and Beta, p = 0.6238; Alpha and Delta p = 0.0957; Beta and Delta, p = 0.1350; Delta and Omicron, p = 0.2408. Alpha variant viral loads (median CT = 27) were lower than in the case of Omicron (median CT = 21) (p = 0.0449) infections; and the same trend was observed for Beta variants (p = 0.0249). Of note, samples containing Omicron variants had the lowest median Ct value (higher viral load) in children.
Vaccinations were a cornerstone of the global COVID-19 response, significantly reducing morbidity and mortality. However, the pandemic’s trajectory varied across countries due to differences in age structures, population immunity, variant circulation, and healthcare infrastructure. Pakistan reported approximately 31,000 COVID-19 deaths up to March 2023 - relatively low given its population size - prompting investigation into possible reasons. Factors including natural immunity, routine vaccination coverage (e.g., BCG, influenza), and the rollout of COVID-19 vaccines have been postulated as possible reasons across different parts of the world (Cocco et al., 2021; Sarfraz et al., 2021; WHO, 2024a; Shrwani et al., 2021).
This study examined the phylogenetic diversity of SARS-CoV-2 variants in Pakistan between 2021 and 2022, a period marked by the dominance of Delta and Omicron variants. We analyzed genome sequences from both vaccinated and unvaccinated individuals and conducted a focused comparison on pediatric cases, who represent nearly half of Pakistan’s 240 million population (GOP, 2024). Our results provide important insight into viral transmission dynamics in a low-resource, high seroprevalence setting.
A key finding was the lack of significant difference in variant distribution between vaccinated and unvaccinated individuals. Despite assumptions that immunological pressure might favor certain variants in vaccinated hosts, we observed similar lineage patterns and genomic divergence across groups. This suggests that during the Delta and Omicron waves, SARS-CoV-2 continued to spread through both immune and non-immune hosts without a marked shift in variant selection.
This aligns with reports of widespread subclinical transmission in Pakistan, including rural and urban areas, likely fueled by high rates of asymptomatic or mild infections (Iqbal et al., 2025). Earlier studies found population-level SARS-CoV-2 seroprevalence reaching 81% by November 2021 (Iqbal et al., 2025), even as testing rates remained low and many infections went undiagnosed. Such high exposure may have buffered the population against severe disease outcomes, regardless of vaccination status.
Importantly, genomic divergence values also showed no significant difference between children (<18 years) and adults, indicating that circulating variants were broadly shared across age groups. Pediatric data are often underrepresented in genomic surveillance, yet children made up 71% of our cohort due to the availability of vaccination status in this group. Our pediatric-focused analysis, both nationally and from a single-center cohort in Karachi, confirmed that Delta dominated in 2021 and Omicron in 2022 - mirroring trends seen globally (Campbell et al., 2021; Kung et al., 2024).
Interestingly, Omicron infections in children were associated with significantly lower Ct values (i.e., higher viral loads) compared to earlier variants, consistent with increased transmissibility (Puhach, Meyer, & Eckerle, 2023). These findings corroborate prior reports in the general population (Nasir et al., 2023) and underscore the role of Omicron in driving large pediatric infection waves, despite lower severity.
A prior study by Abbas et al. (Abbas et al., 2023) showed fluctuating variant dominance in hospitalized children, but did not assess genomic divergence or compare vaccinated vs. unvaccinated hosts. To our knowledge, this is the first study in Pakistan to systematically compare SARS-CoV-2 variant diversity by vaccination status and age group using phylogenetic divergence metrics.
We also observed that Islamabad and Karachi accounted for most sequence submissions during the study period - consistent with their status as international hubs and prior reports on variant introductions into Pakistan (Aziz et al., 2023; Ali, Shah, & Siddiqui, 2020; Bukhari et al., 2023; Nasir et al., 2022). This geographic concentration reflects testing and sequencing infrastructure disparities and highlights the need for broader surveillance coverage across provinces.
This study has several limitations. Vaccination status was not available for many adult cases in GISAID, leading to underrepresentation of this group. Additionally, clinical data such as symptom severity, hospitalization, and comorbidities were unavailable, limiting our ability to correlate viral variants with disease outcomes. Finally, the retrospective design limits causal inferences.
Our findings suggest that in settings with widespread prior exposure, variant distribution and transmission dynamics may not differ significantly by vaccination status - a key consideration for future surveillance and vaccine deployment strategies. The high seroprevalence, young population structure, and mixed vaccine types in Pakistan offer a unique lens through which to understand SARS-CoV-2 evolution in diverse immunological landscapes.
Continued genomic surveillance, particularly in pediatric and rural populations, remains vital to detect emerging variants and guide context-specific public health responses. Moreover, insights from populations with high natural immunity and mixed vaccine platforms can inform global preparedness for future respiratory virus outbreaks.
This study provides novel insights into SARS-CoV-2 variant circulation in Pakistan during the Delta and Omicron waves, highlighting that viral lineages did not differ significantly between vaccinated and unvaccinated individuals, nor between children and adults. Genomic divergence analyses revealed comparable strain diversity across age groups and vaccination status, suggesting widespread community transmission of similar variants irrespective of host immune background.
Our findings underscore the importance of understanding SARS-CoV-2 dynamics in populations with high background immunity - whether from prior infection, vaccination, or both. In such settings, variant-specific susceptibility may be less pronounced, and asymptomatic or mild infections may sustain transmission. This is particularly relevant for low- and middle-income countries where young age demographics, subclinical infections, and mixed vaccine regimens influence viral evolution.
As the pandemic transitioned into an endemic phase, sustained genomic surveillance - especially in pediatric populations and underrepresented regions - is essential to track emerging variants and guide tailored public health interventions. Our results reinforce the need for integrated virologic, immunologic, and demographic approaches to inform global pandemic preparedness in the years ahead.
This study was conducted in accordance with the principles of the Declaration of Helsinki. Ethical approval was obtained from the Ethics Review Committee of Aga Khan University, which granted a waiver of written and verbal informed consent (Approval No. 2021-6316-18156 and 2022-6871-22433) on the grounds that this was a retrospective study using secondary data, for which obtaining consent was impractical. All data was de-identified before retrieval from AKUH database and therefore poses no risk of confidentiality to subjects.
Pre-print version of this article: Mwendwa, F., Ashraf, J., Kanji, A., Bukhari, A. R., Hasan, R., & Hasan, Z. (2025). Temporal Mapping of SARS-CoV-2 Variants During the Pandemic Reveals Similar Genomic Divergence in Unvaccinated and Vaccinated Cohorts, and in Both Adults and Children. Preprints. https://doi.org/10.20944/preprints202504.2203.v1
Supplementary Table 1: Data downloaded from GISAID for samples collected May 2021 to October 2022
Supplementary Table 2: Breakdown of Next clade and Pangolin lineage of the samples collected during the study period, May 2021 to October 2022
Supplementary Table 3: Genomic divergence data
Data is available under the terms of the Creative Commons Attribution 4.0 International.
Sequence data that support the findings of this study have been deposited in the GISAID database, www.gisaid.org and can be accessed at DOI: https://doi.org/10.55876/gis8.250926ct. In addition, underlying data as well as the supplementary data can be found on the Zenodo repository: Genomic Epidemiology of SARS-CoV-2 variants in unvaccinated children found to be similar to that of adults with COVID-19 vaccinations https://doi.org/10.5281/zenodo.17233944.
Table 1. Demographics of the study cohort in the context of SARS-CoV-2 variants
Table 2. Age categories of the individuals in the study as per their vaccination status
Figure 1. Province-wise distribution of samples
Figure 2. SARS-CoV-2 phylogenetic divergence based on vaccination and time of sample collection from May 2021 to October 2022
Figure 3. Age-wise comparison of SARS-CoV-2 variants
Figure 4. Study of SARS-CoV-2 variants in children in the study cohort
We thank David Spiro and Zeba Rasmussen for their guidance and advice during the study.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomic epidemiology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |
|---|---|
| 1 | |
|
Version 1 21 Oct 25 |
read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)