Keywords
Biodiversity, Research data, FAIR, Bioschemas, Harmonization, Mapping, cross-disciplinarity, NFDI
This article is included in the Software and Hardware Engineering gateway.
This article is included in the Reproducible Research Data and Software collection.
This article is included in the Data: Use and Reuse collection.
Bioschemas is a community-driven initiative for enhancing the semantic description and interoperability of life science resources through standardized metadata vocabularies. While initially developed to address needs in diverse life science domains, Bioschemas offers significant untapped potential for improving data harmonization, discoverability, and reusability in fields such as biodiversity research.
This white paper details the implementation of Bioschemas across the NFDI4Biodiversity consortium, a collaborative network within Germany’s National Research Data Infrastructure (NFDI) dedicated to improving findability, accessibility, and reusability of biodiversity data. We outline the motivations and strategic value behind adopting Bioschemas, including on the one hand increased discoverability for a diverse audience—from researchers and policymakers to the public and industry—via semantically enriched, machine-actionable metadata that meets FAIR (Findable, Accessible, Interoperable, Reusable) guidelines. And on the other applying Bioschemas as a unifying, cross-disciplinary metadata approach aimed at improving interoperability within national and European data ecosystems such as the NFDI and EOSC and beyond.
The paper outlines technical workflows, such as the automated transformation of established biodiversity metadata standards (e.g., ABCD) into Bioschemas-compliant representations, and their integration into core infrastructure components, including the consortium’s search and knowledge graph environments. Concrete implementation scenarios, challenges encountered, and harmonization efforts are presented alongside actionable recommendations for technical and non-technical audiences. The paper ultimately provides a transparent account of consortium-internal practices, strategies for community-wide adoption, and perspectives for ongoing development, positioning Bioschemas as a catalyst for open, interoperable, and future-ready biodiversity data infrastructures.
Biodiversity, Research data, FAIR, Bioschemas, Harmonization, Mapping, cross-disciplinarity, NFDI
Bioschemas.org [1] (hereafter referred to as Bioschemas) is based on the markup technology schema.org and its extensions developed by the Bioschemas.org community which were specifically designed for applications in the life-sciences.1 Schema.org [2], which was initiated by major search engine providers including Google, Microsoft, Yahoo, and Yandex, aims to structure web content using controlled vocabularies. This technology enhances internet search results by enabling search engines to better understand page content in a structured, machine readable format.
However, this technology is not limited to the description of website content, but can also be exploited for other web resources. By implementing schema.org markup, datasets gain discoverability through semantically enriched, highly interconnectable metadata, increasing their accessibility to search engines and automated discovery services. As a result, annotated data resources can be prominently featured in search engine results and specialized data portals such as Google Dataset Search [3]. Because data discovery leverages established, robust search infrastructure and aligns with common web search practices, researchers and other interested parties can locate more relevant results with greater efficiency.
However, while schema.org already includes terms related to citations, datasets, and data repositories, it lacks specific terms needed to adequately annotate research entities in the life science domain. Bioschemas addresses this gap by proposing missing concepts (types) and properties to the schema.org community while reusing existing ones where possible. Before new (or complimentary) types and properties are proposed, however, their scope and definitions are thoroughly validated and refined in multiple iterations by the Bioschemas community.
In order to provide consistency of markup within life science communities and identify the essential properties for describing a resource, Bioschemas defines usage profiles over the schema.org types. The Bioschemas profiles provide constraints and guidelines in terms of:
• Marginality - which properties must be used (minimum), which should be used (recommended), and which could be used (optional)
• Cardinality - how many entities can be described (one or many)
• Usage of relevant (e.g., domain-specific) controlled vocabularies and ontologies
Implementing Bioschemas can significantly enhance the quality of data search results. By leveraging controlled vocabularies, search engines gain a deeper understanding of search context, enabling them to deliver more relevant and precise results. In addition to increasing dataset discoverability via general search engines, incorporating schema.org markup into dataset web pages (usually referred to as landing pages) ensures that these datasets are indexed and accurately represented in data search catalogues such as Google Dataset Search.
NFDI4Biodiversity, one of the nine pioneering consortia of the national large-scale research data management and infrastructure project German National Research Data Infrastructure (NFDI), recognizes the potential of Bioschemas to leverage the general visibility, discoverability and integrability of data from the biodiversity domain for a variety of purposes and application scenarios, and has officially committed to using it as its primary discovery standard.
Members of NFDI4Biodiversity use a range of metadata standards to describe their multi-faceted data holdings that serve a wide variety of purposes and functional roles relevant for biodiversity research. A significant proportion of these data holdings are provided in the Access to Biological Collection Data (ABCD) [4] format, an internationally recognized (meta) data standard by the Biodiversity Information Standards group (TDWG).2 ABCD has been adopted within the NFDI4Biodiversity project and its predecessor, the German Federation for Biological Data (GFBio), as a harmonizing standard that supports the exchange of collection and observation data (Type 1, see chapter 2.4 for an introduction to these data types). ABCD is a comprehensive, specialized standard serialized in XML [5], and was developed in the community for the community. Records in ABCD contain dataset level metadata such as dataset title, description, authors and licences, and content information about specific instances of specimens or observations, which are organized in sub-elements of a ABCD dataset called units. Units refer to individual observations and collection objects and can contain details like taxonomic information, measurements, collectors and links to multimedia data. ABCD records thus always contain dataset metadata and unit properties. Accordingly, ABCD records are structurally relatively complex, and its content is not readily accessible beyond specialized, community specific tooling and infrastructures. The latter is also true for other metadata standards used by members of the consortium that are considered similarly specific to certain community needs and purposes.
As a consequence, the adoption of Bioschemas provides a series of advantages in regard of the desired harmonization and convergence of data pools in the consortium, and closes a number of gaps and shortcomings with respect to a broader discoverability, accessibility and integration of biodiversity data for non-academic and cross-disciplinary application scenarios.
Increase findability and interoperability for biodiversity data resources
One of the specific strengths of Bioschemas is its ability to provide a relatively uncomplicated and cost-efficient approach to significantly leverage the discoverability of scientific data resources on the web.3 Implementing Bioschemas for biodiversity data significantly improves interoperability and cross-disciplinary application potential by enabling the use of a common, web-friendly vocabulary that is both machine- and human-readable. Bridging semantic gaps between diverse data sources by providing harmonized, extensible metadata structures, Bioschemas makes datasets not only more discoverable in general and domain-specific search engines but also more readily integrated into broader scientific workflows and digital research infrastructures such as those in development for the NFDI and for the European Open Science Cloud (EOSC). Bioschemas encourages the life sciences community to align key terms and structures, reducing barriers caused by heterogeneous metadata practices and enabling seamless aggregation, comparison, and reuse of data across related disciplines, including ecology, genomics, and environmental sciences. Ultimately, this standardization facilitates interdisciplinary analyses, supports FAIR principles (Findable, Accessible, Interoperable, Reusable), and lowers the technical threshold for data sharing and synthesis at scale.
Improving the FAIRness of biodiversity data
The FAIR principles provide a widely adopted framework for effective research data management,4 ensuring that data can be easily discovered, accessed, integrated, and reused by both humans and machines. Adhering to these principles enhances research transparency, reproducibility, and the overall value of scientific data.
Web services for assessing the FAIRness of data, such as the FAIR-checker service from the French Institute for Bioinformatics5 and the automated FAIR data assessment tool “F-UJI” from the FAIRsFAIR project,6 promote the use of schema.org markup. In combination with complementary web technologies such as HTTP Signposting [6] and Content Negotiation [7] it helps ensure that annotated data resources are more easily navigable and interpretable by automated systems.
An adoption of Bioschemas and these web technologies will consequently increase the FAIRness of NFDI4Biodiversity data, and reward individual efforts in that regard with measurably higher Fairness scores.
Make knowledge about biodiversity available in knowledge graphs
Bioschemas, being fully compatible with W3C RDF [8] and Linked Data specifications, provides a robust foundation for representing biodiversity data in a structured and interconnected manner. Using JSON-LD [9], the current recommended serialization format by schema.org, data described with Bioschemas can be seamlessly integrated into knowledge-graph-based infrastructures. This integration facilitates the cross-linking of various biodiversity resources, enabling the creation of rich, interconnected datasets. The use of Bioschemas enhances data discoverability and analysis by supporting complex semantic queries on annotated biodiversity datasets, allowing researchers to uncover relationships, patterns and insights that were previously hidden in isolated silos.
Improved discovery and integrability of data in NFDI4Biodiversity infrastructures
The effective discovery and integrability of data across NFDI4Biodiversity infrastructures critically depends on the ability to unify and harmonize diverse metadata sources. Thus far, full connectivity has primarily been limited to data described in the ABCD format and selected PANGAEA holdings, leaving other prevalent schemas (such as EML, DwC-A, and SDD) [10][11][12] only partially integrated with central services like the search stack and related infrastructure components. This situation not only mirrors the consortium’s heterogeneous data landscape but also underscores the inherent constraints of existing standards.
ABCD, as the backbone for specimen and occurrence data (Type 1), represents a well-established but highly specialized schema that is not easily extensible to a broader range of data types and scientific questions addressed in biodiversity research. Its complex structure limits its applicability when describing, for example, taxonomic, environmental, analytical, or sequence data. Consequently, a more general and flexible standard is indispensable to support the full range of functional and disciplinary requirements of the consortium.
The adoption of Bioschemas meets this need by providing a unified, web-native framework for semantically enriching and harmonizing metadata across all relevant data types. By establishing Bioschemas as a cross-cutting discovery layer, NFDI4Biodiversity can unlock seamless, central access to distributed data resources via core services — most notably the central search portal (GFBio Search), knowledge graph systems, and future-oriented infrastructures like the Research Data Commons (RDC) [13], and relevant components such as the terminology service BiodivPortal [14]. This architecture allows the aggregation and indexing of heterogeneous metadata, making diverse datasets accessible and interoperable not only within, but also beyond, the consortium.
Transitioning core systems to Bioschemas as outlined in subsequent chapters streamlines and future-proofs the aggregation and discovery of research assets, supports more consistent semantic integration, and lowers barriers for new data types and technology adoption. Integrating Bioschemas tightly into infrastructural workflows will thus provide the necessary foundation for comprehensive cross-resource search, knowledge graph creation, and interoperability with external initiatives.
The adoption of Bioschemas as a central standard for discovery metadata has been a key focus within NFDI4Biodiversity. The consortium has formally endorsed Bioschemas as the preferred unifying standard for discovery metadata at the NFDI4Biodiversity All Hands Conference in Berlin in 2022 and has developed important foundations for an adoption of Bioschemas through collaborative efforts. Building on this consensus, these efforts and activities will expand and concretize in accordance with the implementation strategies outlined in the following sections.
Services for the consortium and the community
The adoption of Bioschemas in NFDI4Biodiversity Data Centers and other relevant data providers will be supported by a series of activities:
Implementation guidelines:
Comprehensive guidelines will be provided for implementing Bioschemas, outlining best practices and technical requirements to ensure successful integration across diverse systems.
Training and community engagement:
Targeted training sessions and discussion forums will be organized to educate stakeholders on the benefits and technical aspects of Bioschemas. These activities will also create platforms for feedback and knowledge sharing, fostering a collaborative community around Bioschemas adoption.
Transformation pipeline service:
A robust transformation pipeline service will be developed to enable the creation of Bioschemas representations for data assets serialized with major NFDI4Biodiversity consensus content standards such as ABCD. This service aims to streamline data integration from various sources, promote interoperability, and significantly enhance the discoverability of biodiversity data.
Data integration into knowledge graph infrastructures
NFDI4Biodiversity will drive the implementation of Bioschemas to support a wide range of use-cases with respect to Knowledge Graph infrastructures, including but not limited to respective activities in NFDI such as KG4NFDI [15].
Integration pipeline for Bioschemas markup
In support of these activities, NFDI4Biodiversity will develop and implement an integration pipeline that enables the efficient processing of NFDI4Biodiversity-related Bioschemas markup for emerging Knowledge Graph infrastructures in the NFDI and beyond. This pipeline will support the integration of annotated data into Knowledge Graphs, promoting data interoperability and facilitating the discovery and reuse of biodiversity data within and across disciplinary boundaries.
Enhanced BiodivPortal Annotator service
The BiodivPortal Annotator [16] is a tool to enhance text by adding suitable annotations to semantic concepts related to terms used in the text. This helps to avoid confusion caused by homonyms and increases the comprehensiveness and the ability to process the texts for, for example, search matching or semantic reasoning. The consortium will adapt and extend the BiodivPortal Annotator service to incorporate standard terminology URIs within Bioschemas annotations. These developments will enable the accurate and consistent application of standardized terminology, thereby improving the quality and interoperability of annotated data.
Mappings and crosswalks for interoperability
To facilitate data exchange and integration across different systems, NFDI4Biodiversity will establish mappings and crosswalks between Bioschemas and other standards such as ABCD and Ecological Trait-data Standard (ETS),7 available in a Semantic Web format. These mappings will enable the conversion of (meta) data between different formats, promoting data interoperability and facilitating the discovery and reuse of biodiversity data.
Collaboration with related communities and consortia
In alignment with the One NFDI vision, NFDI4Biodiversity is actively fostering collaboration with related scientific communities and their representative NFDI consortia to facilitate the discovery and exchange of knowledge and data resources.
Interoperability efforts with FAIRAgro [17] and DataPLANT [18]
NFDI4Biodiversity has initiated an ongoing exchange with the other NFDI consortia FAIRAgro and DataPLANT to harmonize the implementation and use of schema.org markup technologies, with a focus on:
1. Terminology Harmonization: Utilizing similar or compatible terminologies to ensure consistency and accuracy in data annotation.
2. Markup Syntax Harmonization: Standardizing markup syntax to enable seamless data exchange and integration.
3. Data Granularity Harmonization: Harmonizing typical data granularities to ensure compatibility and interoperability across different systems.
Expanding interoperability efforts
While NFDI4Biodiversity plans to intensify and consolidate the exchange and harmonization efforts in the coming years, the natural next steps is to extend collaborations to other relevant NFDI consortia such as NFDI4Microbiota and NFDI4Earth in order to promote broader data interoperability and exchange with this topically closely related communities.
NFDI4Biodiversity, in close collaboration with other NFDI activities and groups—such as the Section Metadata, and other consortia—will leverage Bioschemas to significantly enhance the discoverability and accessibility of its data pools. By promoting Bioschemas as one of the primary standards for data discovery within the NFDI,8 the consortium aims to foster greater interoperability and strengthen the overarching research data infrastructure. Furthermore, the reuse of schema.org types and properties by other NFDI consortia facilitates seamless interoperability across scientific domains,9 supporting collaboration and enabling the integration of biodiversity data with datasets from other disciplines.
The adoption and implementation of Bioschemas is gaining momentum not only within national initiatives but also across major European and international biodiversity research infrastructures. ELIXIR [19], as a leading European life sciences infrastructure, actively supports the use of Bioschemas in biodiversity-related use cases, as reflected in dedicated working groups and implementation studies. ELIXIR’s collaborations span a wide ecosystem of projects and infrastructures, including BiCIKL (Biodiversity Community Integrated Knowledge Library), Biodiversity Genomics Europe (BGE), BioDT (Biodiversity Digital Twin), and national initiatives like ARISE (Netherlands), e-BioDiv (Switzerland), and the Pole National de Données de Biodiversité (France).10 These efforts focus on harmonizing metadata standards, improving data interoperability, and connecting molecular and biodiversity data across borders and disciplines.
Globally, major infrastructures such as the Global Biodiversity Information Facility (GBIF) and EMBL-EBI’s BioSamples database are exploring or implementing Bioschemas and related web standards to enhance data discoverability and integration [20].11 Together, these efforts illustrate an international movement towards harmonized, machine-readable metadata formats—with Bioschemas acting as a catalyst for improving scientific data findability, accessibility, and interoperability across life science and biodiversity data domains. By aligning with this global movement, NFDI4Biodiversity ensures that its efforts are compatible with, and contribute to the global network of biodiversity data resources. This alignment not only accelerates scientific discovery and cross-disciplinary collaboration but also supports evidence-based policy and conservation efforts at both European and international levels.
This chapter provides a comprehensive foundation for understanding the strategic adoption and implementation of Bioschemas within the NFDI4Biodiversity consortium. It highlights the central role of the consortium’s technical infrastructure and supporting services (illustrated in Figure 1) in facilitating this transformation process, detailing key considerations for the generation, storage, and management of Bioschemas metadata across distributed systems. We place particular emphasis on the detailed description of the mechanisms of centralised data search and aggregation in the consortium, and highlight the implications, benefits and challenges of a broad adoption of Bioschemas for this key infrastructural component. The chapter further situates the discussion of diverse biodiversity data types within the context of advancing data discoverability, interoperability, and notably cross-disciplinary integration—core objectives that underpin the consortium’s efforts to harmonize metadata across heterogeneous research outputs. It outlines how Bioschemas serves as a unifying semantic framework, enabling the effective transformation and enrichment of established domain-specific data formats into machine-actionable representations that amplify the visibility and reuse potential of biodiversity resources. Together, these elements form a cohesive narrative that shapes the roadmap for ongoing and future endeavors to embed Bioschemas as a primary discovery metadata standard, driving forward NFDI4Biodiversity’s vision of an integrated, FAIR-aligned biodiversity data ecosystem.
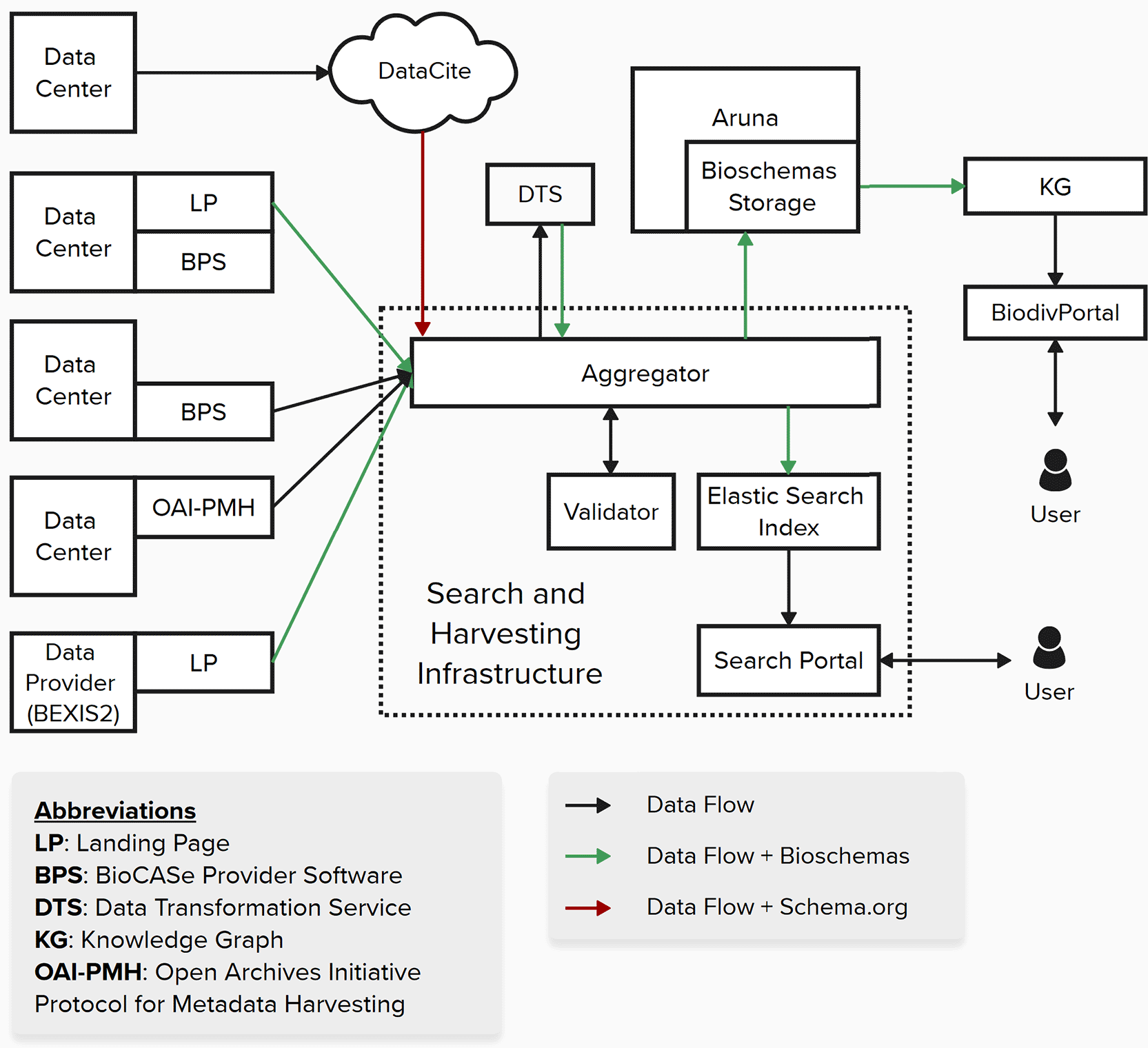
The NFDI4Biodiversity vision of the interplay of relevant infrastructure components, supporting services and interfaces for providing, aggregating, processing and disseminating Bioschemas representations.
The implementation of Bioschemas will involve various components of the NFDI4Biodiversity infrastructure. Each of these components will play a specific role in generating, processing, or utilizing Bioschemas markup to enhance data discovery and integration across the consortium and beyond.
GFBio data centers
The GFBio Data Centers [21] represent dedicated institutional partners within NFDI4Biodiversity, entrusted with the curation, management, archiving, and publication of diverse research datasets spanning the major data types described in Section 2.4. The group currently comprises 10 Data Centers, 7 of which serve specifically as Collection Data Centers, publishing specimen and occurrence data using the ABCD standard. These vast data pools are harvested and made centrally searchable by the GFBio Search Stack. By centralizing and harmonizing data provision, these centers act as a foundational pillar for unified data discovery and access to biodiversity data within the consortium.
BioCASe provider software
The BioCASe Provider Software (BPS) [22] serves as a flexible middleware solution for publishing data from relational databases into broader information networks. Although inherently format-agnostic, BPS is operated by most Collection Data Centers within NFDI4Biodiversity to expose specimen and observation data using the ABCD standard. During the initial setup, institutions configure BPS by mapping database tables and columns to corresponding ABCD elements. Once this mapping is established, data can be disseminated either through the export of ABCD archive files or via dynamic, on-demand exchanges utilizing the BioCASe Protocol. Within the consortium’s infrastructure, the GFBio Search and Harvesting Stack relies exclusively on these ABCD archives for data ingestion and further processing.
Research data commons: Aruna
ARUNA [23] is a performant and geo-redundant data storage solution integrated within the RDC (Research Data Commons) environment. It offers advanced metadata management compliant with the FAIR data principles and supports hierarchical organization by grouping digital assets into projects, collections, datasets, and objects. Aruna facilitates flexible linkage between entities, allowing detailed specification of relationships and versioning. The platform supports attaching multiple metadata objects to each dataset, enabling concurrent annotation through standards such as ABCD and Bioschemas. Moreover, Aruna’s orchestration features—such as web hooks—enable automated pipeline actions, including seamless transformation of ABCD metadata to Bioschemas representations via the Data Transformation Service.
BEXIS2
BEXIS2 [24] is a modular, and web-based research data management system designed for use across scientific domains such as ecology, biodiversity, and soil science, and is adaptable to other disciplines.12 BEXIS2 instances typically host structured research data (tabular datasets) along with rich metadata and supplementary files. During the setup or configuration of a BEXIS2 instance, an administrator can define how individual metadata fields or complete metadata sets are mapped to the Bioschemas Dataset Profile using an integrated mapping tool. This built-in mapping mechanism improves the web discoverability of datasets through semantic markup and demonstrates how existing research data management systems can be enhanced to support Bioschemas. Additionally, BEXIS2 supports biodiversity data exchange through Darwin Core Archive exports and can facilitate data preparation for submission to data publication infrastructures such as GFBio and PANGAEA.
Data transformation service
The Data Transformation Service (DTS) [25] delivers essential functionality for data interoperability, supporting conversion between multiple community standards in biodiversity informatics. As a web service, DTS enables both batch and on-demand transformations of formats such as ABCD and DwC, allowing Data Centers and providers to efficiently generate Bioschemas-conformant representations from community formats. The service offers version control, OpenAPI documentation for transparent integration, and temporary result retention for asynchronous consumption workflows. The latest enhancements include first drafts of ABCD-to-Bioschemas conversion, supporting seamless integration of discovery metadata across NFDI4Biodiversity’s distributed infrastructure.
Diversity workbench
The Diversity Workbench [26] is an open-source, modular management system enabling structured curation and publication of bio- and geodiversity data. It supports robust data pipelines, allowing datasets to be published using standards like ABCD, with BioCASe Provider Software providing the necessary export functionality. For Type 2 and Type 3 data, additional open-source tools (such as DTN REST web services and DiversityNaviKey) extend the platform’s publication and annotation capabilities,13 facilitating semantic enrichment and interoperability with Bioschemas and associated standards.
BiodivPortal and the NFDI4Biodiversity knowledge graph
The BiodivPortal [27] is a specialized ontology repository and semantic service platform integral to the NFDI4Biodiversity infrastructure. As a core consortium service, it maintains a curated, FAIR-compliant catalogue of terminologies, ontologies, and controlled vocabularies tailored to the biodiversity and environmental sciences, providing essential resources for semantic data annotation and integration. The portal serves as the backbone for the semantic management of data, supporting the creation and maintenance of the NFDI4Biodiversity Knowledge Graph, and supplying community-driven tools for harmonizing, annotating, and linking trait and observation data across heterogeneous sources. Additionally, BioDivPortal offers collaborative and technical support, enabling versioning, provenance tracking, and community enrichment of semantic resources, which together ensure interoperability, advanced searchability, and data FAIRification throughout the consortium’s research infrastructure.14
The GFBio search and harvesting infrastructure is a modular ETL (Extract - Transform - Load) pipeline. It collects, processes, and indexes biological research data from the official GFBio data centers, the European Nucleotide Archive (ENA), and professional societies, consolidating them into a unified, searchable repository powered by an Elasticsearch index. The stack is planned to be involved in driving the data integration and discovery within the NFDI4Biodiversity RDC environment, enabling researchers in the domain to discover and access diverse biological datasets through a centralized search portal [28]. In the following sections we introduce the core components of the search and harvesting infrastructure, explain a typical flow of data before we dive into how the architecture may benefit from using bioschemas.
Core components
1. Aggregator: Serves as the entry point where data providers register and manage their institutional and project-based datasets through a web interface. It offers an API that other components, particularly the harvesters, use to access registered datasets.
2. Validator: Used by the aggregator to validate registered datasets. Currently supports the ABCD standard, checking for general schema validity and compliance with GFBio data center metadata consensus elements [29]. Feeds validation results back to the aggregator, providing feedback to users during data registration about validity and conformance levels.
3. ABCD Harvester: Connects to the aggregator to retrieve all registered datasets, downloads ABCD archives, and extracts XML files for processing. The harvester then processes these files using resource-efficient streaming techniques, transforms ABCD data into a standardized format suitable for indexing, and pushes the transformed data into the index.
4. Portal Harvester and Index: Manages the Elasticsearch index (e.g. Aliases) and provides interfaces for harvesting various formats beyond ABCD (e.g., via OAI-PMH [30], DataCite [31]). Connects to multiple data sources, retrieves metadata, and transforms it into the index’s target format PanSimple [32]. Similar to the ABCD harvesting process, it processes the retrieved metadata and pushes the transformed data into the Elasticsearch index.
5. Search-UI: A user-friendly web interface that enables researchers to discover and access biological datasets through the Elasticsearch index. Features include faceted simple and advanced searches, rich metadata previews with dataset landing page links, geographic visualization, and taxonomic browsing.
Data flow
Currently, data providers register and validate their datasets in the Aggregator. The Data Downloader retrieves these datasets, which are then processed by the ABCD Harvester and indexed into Elasticsearch by the Portal Harvester. The Portal Harvester also features adapters that enable direct harvesting from DataCite or OAI-PMH endpoints [33], providing flexible setup options for harvesting from a wide variety of providers. Through this pipeline, users can ultimately search and access the aggregated data via the GFBio search interface. This architecture provides a flexible foundation that would strongly benefit from an extension to support schema.org and Bioschemas standards.
Enhancing the system with Bioschemas
The GFBio search and harvesting architecture presents an ideal case for implementing schema.org and Bioschemas markup throughout its entire workflow. As a complete data pipeline, from initial registration through harvesting and indexing to final discovery, the system would significantly benefit from adopting standardized vocabularies across all components. This standardization would enhance data interoperability, improve metadata quality, and facilitate more effective data discovery. The three-layer architecture provides natural integration points for Bioschemas. The following sections detail how each layer would benefit from Bioschemas integration.
Source layer: Enhancing data provider content
The Aggregator holds a central position in the system, serving not only as the registration point but also providing an API for catalog consumption. Beyond these core functions, it can orchestrate various services around registered data, including transformations, validations, and statistical analysis. While the data catalog itself may utilize standardized vocabularies like DCAT [34] (Data Catalog Vocabulary), the Aggregator can facilitate Bioschemas adoption by offering transformation services. For known formats, it can delegate to transformation services in the background, converting existing metadata into Bioschemas-compliant representations. By positioning the Aggregator as both a catalog and orchestration hub of dataset related functionality, it can help the network gradually transition to standardized Bioschemas descriptions while supporting providers at various stages of adoption.
Validation and harvesting layer: Enriched data collection
The validation and harvesting components work in tandem to ensure data quality and standardization. Currently, validation checks data against XML schemas and for GFBio Data Center rules before datasets are harvested. With Bioschemas integration, both layers will be enhanced. The validation component will extend its checks to include Bioschemas compliance, verifying JSON-LD structure and required properties. For datasets in compatible formats but lacking Bioschemas markup, the harvesting layer could also invoke the respective DTS pipelines to generate compliant metadata during the processing. This ensures all data, whether natively in Bioschemas or transformed, meets the same quality standards before indexing.
Index layer: Discovery infrastructure
The search index would benefit significantly from adopting Bioschemas as the target format for all data sources, replacing the current PanSimple format. Using schema.org plus Bioschemas as the canonical target in the ETL pipeline means every source maps once to a single, well-established vocabulary already understood by search engines and life-science harvesters. This “write once, reuse everywhere” approach delivers multiple advantages: it eliminates redundant mapping overhead, maintains human-friendly field names for UI facets, and ensures the system remains future-proof through its modular design. Furthermore, standardizing on Bioschemas can drive alignment efforts and facilitate cross-consortia harmonization for data discovery within the NFDI ecosystem.
Conclusion: Benefits and strategic implications
The integration of Bioschemas throughout the GFBio search and harvesting stack exemplifies the practical implementation of the NFDI4Biodiversity consortium’s commitment to standardized discovery metadata. By adopting Bioschemas across all three architectural layers from data registration through processing to discovery, the system demonstrates how existing biodiversity infrastructure can evolve to meet modern interoperability requirements.
This architectural transformation positions the GFBio stack as a reference implementation for other NFDI4Biodiversity components and demonstrates the feasibility of consortium-wide Bioschemas adoption. As outlined in sections 1.3 and 2.6, this approach supports the broader vision of cross-consortia data discovery and positions biodiversity data within the emerging web of FAIR scientific resources.
The generation and storage of Bioschemas representations within NFDI4Biodiversity infrastructure are streamlined to maximize consistency, efficiency, and findability across diverse data providers and system architectures. Bioschemas markup for datasets or individual unit-level data can be created at two main points in the data flow: either directly by the Data Centers and Data Providers responsible for the original research data, or automatically by the Aggregator (or central harvesting system) through the Data Transformation Service (DTS) when datasets are registered and processed during the harvesting workflow.
The preferred approach is for providers to transform and generate Bioschemas markup at the source, as part of their publication or export processes; this ensures that the semantic description closely matches both the original metadata source and its updates, and can be embedded directly into dataset or unit landing pages for optimal machine-readability and discoverability. When native provision at this level is not feasible—due to technical or resource limitations—the centralized DTS provides a fallback, enabling batch or on-demand transformation of established community formats (e.g., ABCD, DwC-A) into Bioschemas-compliant JSON-LD, which can then be harvested and indexed across the consortium’s infrastructure.
To maintain high searchability and seamless integration with external data discovery services, Bioschemas markup should be generated at publication time and stored alongside the source dataset files—ideally as static JSON-LD files or as embedded markup within landing pages. This approach avoids delays that would occur with on-the-fly transformation and ensures that updates to underlying datasets prompt corresponding updates to the Bioschemas representations. Therefore, workflows should ensure Bioschemas files are refreshed whenever the dataset changes, or as part of regular maintenance cycles. It is not recommended to embed dynamically generated markup into HTML via JavaScript after user-page loads, as this prevents effective crawling by search engines and automated ingest systems.
If a dataset or unit is withdrawn or unpublished, the associated landing page should persist as a “tombstone page” that continues to serve the corresponding Bioschemas metadata, clearly indicating the dataset’s retraction status and retaining its discoverability for provenance and citation purposes. This necessitates the permanent, protected storage of Bioschemas files even after data removal, with suitable mechanisms to prevent accidental overwriting during routine updates. This ensures the robustness and longevity of machine-actionable discovery metadata throughout the entire NFDI4Biodiversity lifecycle.
The NFDI4Biodiversity consortium encompasses a broad spectrum of research institutions and data providers, each contributing diverse types of biodiversity and environmental data oriented towards complementary scientific purposes (see Table 1). These data resources are categorized following the GFBio “Major Types of Biological Data” [35], with each category presenting unique formatting and metadata requirements. An overview of the main data types and commonly used community standards is provided below:
• Type 1: Biodiversity and occurrence data
Specimen and observation data documenting the physical presence or recorded instances of organisms (e.g., herbarium collections, faunal surveys). These datasets are predominantly provided in the ABCD (Access to Biological Collection Data) standard (XML serialization), as agreed by most data providers in the consortium, facilitating robust exchange and harmonization.
• Type 2: Taxon level data
Taxonomic datasets containing information on names, ranks, synonymy, and classification systems. Formats include DwC-A supplemented with EML and CDM Light [36], ensuring compatibility with nomenclatural services and integrating with broader taxonomic databases.
• Type 3: Environmental, biological, and ecological data
Representing tabular or structured datasets derived from ecological studies, such as long-term ecosystem monitoring, trait assessments, or environmental gradients. Principal formats are EML, DELTA [37], and SDD, with frequent use of spreadsheets and custom database exports, enabling flexible representation of site-based measurements and experimental variables.
• Type 4: Non-molecular analytical data
Datasets resulting from analytical procedures not involving molecular/genomic sequences, such as morphological, chemical, or physical analyses. These may be described using EML or proprietary lab formats, with DataCite [38] often used for dataset-level citation and persistence.
• Type 5: Molecular sequence data
Genomic, transcriptomic, and molecular marker data, including DNA barcodes and sequence-based organismal assignments. Leading formats are MIxS [39], standard sequence formats like FASTA or FASTQ, and GenBank flat files, supporting integration into molecular databases and facilitating downstream genetic analyses.
These categories are based on the functional role each data type serves in biodiversity science: recording existence (Type 1), organizing and referencing biological names (Type 2), providing context through ecological and environmental data (Type 3), supporting analytical interpretation (Type 4), and integrating molecular information (Type 5).
GFBio consensus elements
The group of Data Centers contributing to GFBio, the predecessor project of NFDI4Biodiversity, collaboratively defined a standardized set of core metadata fields required for publishing and exchanging biological and biodiversity data within the project and related infrastructures for Type 1 data provided in ABCD, the “GFBio consensus elements” [40]. These elements ensure that all datasets in the ABCD 2.06 format include essential information such as dataset identifiers, titles, descriptions, authorship, licenses, contacts, and key specimen or observation properties. By mandating these fields, the consensus elements support consistent, high-quality metadata, facilitating interoperability, reliable discovery, and downstream integration of Type 1 biodiversity datasets across diverse related platforms. NFDI4Biodiversity adopted and continued the use of these consensus elements for respective data sources.
To fully realize the benefits of standardized, machine-actionable metadata and enhance the discoverability and interoperability of as much of the consortium’s data holdings as possible, the transformation of primary and secondary data formats into Bioschemas representations is a key technical and strategic objective.
In the following, we outline reasonable scenarios for each major data type, illustrating how data can (or could) be mapped, enriched, and published using Bioschemas profiles and vocabulary extensions.
Please note that a few of the concepts described in these scenarios have been identified as currently missing in the recent Bioschemas profiles and specifications. These instances are clearly marked by italic letters and a trailing asterisk, for example Observation* or Specimen*. We discuss the issue of missing data types and properties in the section 4.4 Challenges and Shortcomings.
Type 1: Biodiversity and occurrence data
Scenario: ABCD records for specimens and site-based observations can be transformed into Bioschemas-compliant “BioSample”, or “Observation” entities, respectively. Providers generate standard-compliant, harmonized Bioschemas representations using the centralized transformation pipelines when they publish or update their datasets. Key attributes, such as taxon names, persistent identifiers, information about the collection* events, or information about time, provenance or the specimen* are rendered in JSON-LD and stored alongside the dataset files on dataset and unit level, allowing direct inclusion in dataset landing pages for machine-readability and automated ingest of discovery services such as Google Dataset Search. In addition to these key components, a rich set of additional ABCD elements transformed to Bioschemas provide the semantic depth to enable enhanced search capabilities and discoverability.
Some of them require suitable abstractions concerning for example spatial coverage or taxonomic information compiling individual occurrence descriptions in ABCD:Units to meaningful generalized representations (for example by clustering lower taxonomic levels).
The transformation pipeline is provided by the Data Transformation Service (DTS) and supports both batch and on-the-fly processing. The latter allows for on-the-fly conversion as a fallback solution for data harvesting and indexing, if on-premise generation shouldn’t be possible. That ensures that all data holdings are represented in the central NFDI4Biodiversity infrastructures such as the Search stack.
Current state of adoption: Since NFDI4Biodiversity Type 1 data providers agreed to provide their holdings in ABCD format compliant to the GFBIO consensus elements, a large proportion of data in NFDI4Biodiversity are available in ABCD format. While being well standardized and widely adopted in the international biodiversity research community, cross-disciplinary provision of metainformation and contents remain a major challenge due the domain-specific nature and complex structure of ABCD records. The adoption of Bioschemas is expected to fill this important gap by providing an additional discovery layer that promises to leverage the findability and assessability of such data and make them available for other user groups and application scenarios. Accordingly, much of the focus of NFDI4Biodiversity has been concentrated on the development of an ABCD - Bioschemas mapping and its application in a transformation pipeline based on the DTS.
In its current state, the DTS provides ABCD-to-Bioschemas file transformations allowing dataset search and discovery based on the ABCD GFBio consensus elements. However, by providing first additional context elements such as taxonomic terms or details about complementary measurements on dataset level, the resulting Bioschemas representations already go beyond that point. The ultimate aspiration in that regard is to transform as much valuable context from ABCD data as possible to enable enhanced search and interoperability scenarios. The recently concluded mapping of single ABCD:Units to Bioschemas “BioSample” type representations are expected to be included in the DTS pipelines as the next step, and mark another important milestone towards reaching NFDI4Biodiversity’s Bioschemas implementation goals.
Type 2: Taxon level data
Scenario: Taxonomic concepts exported in DwC-A+EML or CDM Light are mapped to Bioschemas “Taxon” and “TaxonName” profiles, capturing taxon hierarchy, synonyms, nomenclatural status, and cross-references. Landing pages for taxonomic objects or API endpoints provide JSON-LD markup, supporting semantic search and interoperability with global name registries.
Current state of adoption: Taxon information is already being provided in Bioschemas serialization by a series of early adopter institutions in NFDI4Biodiversity such as the DSMZ, the BGBM (see chapter 3 for details), Senckenberg – Leibniz Institution for Biodiversity and Earth System Research and the IPK - Leibniz Institute of Plant Genetics and Crop Plant Research (both not represented in this document). If the need for syntactical and semantic harmonization should arise among partner institutions, the consortium will offer a forum for discussions to reach a consortial consent.
Type 3: Environmental, biological, and ecological data
Scenario: Measurement-based and site data in EML, DELTA, or SDD are transformed into Bioschemas “Dataset” profiles, with the “variableMeasured” and “PropertyValue” extensions capturing detailed experimental or observational attributes. Associated data files become “DataDownload” entities, referenced within the markup to facilitate automated harvesting and cross-disciplinary linking (e.g., for trait-based or time-series analyses).
Current state of adoption: Type 3 data have not been in particular focus for the Bioschemas adoption by NFDI4Biodiversity yet. However, the provision and distribution of environmental and ecological context information in schema.org is common practice by many Earth & Environmental data providers. PANGAEA for example, provides much of its metadata that is rooted in the geospatial domain and in the ISO19115 standard encoded in schema.org compliant JSON-LD.18 The EML standard is conceptually and structurally very similar to the ISO19115 family of standards, and is subject to international mapping and cross-walk activities also including Bioschemas/schema.org.19
Consequently, EML is considered a manageable challenge concerning the transformation to Bioschemas. However, the adoption and harmonized implementation of Bioschemas for EML data providers remains an objective for the future.
Type 4: Non-molecular analytical data
Scenario: Analytical results, including lab-derived measurements and characterizations, are mapped to Bioschemas “Dataset” with linked “PropertyValue” instances, and metadata enhanced through controlled terms for analytical techniques. Integration ensures both core and derived data are indexed and retrievable within knowledge graphs and search portals.
Current state of adoption: Type 4 data has not been officially addressed in the consortium yet. However, if EML is used as a standard for that category, similar assumptions and objectives as for Type 3 data should apply.
Type 5: Molecular sequence data
Scenario: Sequence submissions in MIxS/FASTA formats are described as “Dataset” entities, with explicit linkage to sample information (“BioSample”), taxon associations (“Taxon”), and protocol metadata (“LabProtocol”). Mappings accommodate accession numbers, sequence attributes, and provenance, supporting both discoverability in molecular repositories and connection to specimen and observation records.
Current state of adoption: The need for a mapping/crosswalk of molecular sequence metadata to schema.org/Bioschemas is acknowledged in community discussions [41], but no official mapping or crosswalk exists to date. Incentivizing such developments is a potential future objective for NFDI4Biodiversity.
Cross-category and mixed data
Scenario: Larger datasets integrating multiple data types (e.g., ecological time series with taxonomic and molecular annotations) deploy composite Bioschemas markup, linking “Dataset”, “BioSample”, “Observation”, and “Taxon” entities. This facilitates advanced search scenarios and seamless reuse in interdisciplinary and trans-consortial research.
Current state of adoption: Data compilations of that category will greatly benefit from the ongoing efforts in NFDI4Biodiversity to represent Type 1 to 5 data in Bioschemas.
This chapter presents a collection of practical implementation examples from NFDI4Biodiversity partner institutions, illustrating how Bioschemas standards are being applied within diverse organizational settings. Each section aims to highlight the specific roles these partners play in the consortium, the types of biodiversity data they manage, the software tools and workflows in use, and the adopted strategies for generating and integrating Bioschemas-compliant metadata. By showcasing these institutional approaches and technical solutions, the chapter provides concrete insights to support broader adoption, foster cross-institutional learning, and demonstrate the tangible benefits and challenges associated with the consortium’s harmonization efforts.
The Botanic Garden and Botanical Museum Berlin (BGBM) has been a GFBio Data Center since the first phase of GFBio. It primarily manages specimen and occurrence data (Type 1), as well as taxonomic data (Type 2), with a specific focus on botanical research. Multiple software products are used to provide access to these research outputs, offering interfaces for entire datasets and individual units such as herbarium specimens and taxon names. Access is provided via human-readable landing pages as well as various machine-readable formats.
Enhancement of these services with Bioschemas markup is planned, prioritizing implementation on dataset landing pages within the NFDI4Biodiversity framework. This includes extending the number of formats supported through HTTP content negotiation.
The Bioschemas types and profiles most relevant to BGBM include Dataset, BioSample (for herbarium specimens), Observation, as well as Taxon & TaxonName. The generation and storage of Bioschemas markup vary across systems; in most cases, it is produced dynamically through direct transformation from internal databases. In some instances, BGBM relies on the Data Transformation Service to convert metadata for dataset landing pages, storing the resulting Bioschemas representations locally.
As a biological resource center, the DSMZ is one of the largest collections of microorganisms and cell lines worldwide. The collection includes bacterial strains, human and animal cell lines, fungal strains, plant viruses and bacteriophages for research purposes. In addition to the physically provided resources, the DSMZ also manages data about these microorganisms and cell lines (usually of Type 2-5) in public scientific databases like BacDive, CellDive and PhageDive,20–22 amended with biological knowledge from different resources.
Schema.org and Bioschemas are currently used in the context of scientific databases at the DSMZ. Every entity in those databases has its own page, in which schema markup is used to leverage the findability of this resource through general search services such as Google or Bing.
Based on the structural philosophy of schema.org, the DSMZ and partners are working on a standardized data model for storing and exchanging microbial strain data. By integrating partial classes from schema.org and Bioschemas in the data model, a quick extraction of valid schema markup with little to no data transformation is the goal.
At the DSMZ the Bioschemas data for every entry of a scientific database is built depending on the respective resource. In BacDive the schema describes the whole dataset and is therefore flagged as a dataset. A BacDive dataset is about one microbial strain. Included in the schema are information of the DSMZ as the providing organization, BacDive as the catalogue, identifiers for the dataset, the link for the described BacDive entity, the license and some keywords about biological properties of the strain. From Bioschemas the taxon class is used to describe the respective species of the strain. Furthermore, Bioschemas are also used for growth media description in MediaDive, another database of the DSMZ.23 The Bioschemas element LabProtocol is used to describe the preparation of specific growth media, including reagentUsed and steps.
At the moment schema.org representations for the dataset websites of our scientific databases are generated dynamically on-the-fly from the datasets, and not stored. The describing schema.org is delivered as a JSON-LD block inside a script-tag within the HTML header.
For example, the DSMZ holds data on the growth conditions required by each microorganism, including temperature, pH level and oxygen tolerance. This data could be converted into the LabProtocol structure of Bioschemas.
PANGAEA Data Publisher for Earth & Environmental Science18 is one of the NFDI4Biodiversity Data Centers hosting data from a wide range of disciplines of the Earth and Environmental Science domain including Biodiversity information. Data publications relevant to Biodiversity research mostly entail occurrence and environmental data (Type 1 & 3), some including taxonomy level considerations (Type 2), and to some extent non-molecular studies (Type 4).
Since PANGAEA’s generalizing approach to the metadata description of the published data focuses primarily on a clear spatial and temporal reference, the in-house metadata schema is primarily based on the geographical content standard ISO19115, but has also been delivered in JSON-LD serialized schema.org alongside other export formats for many years. The meta information for data records to be described with schema.org is integrated into the HTML source code of their respective landing pages with the <script> element, but is also made available as metadata records that can be accessed directly using FAIR signposting [42] and HTTP content negotiation [43] technologies.
In addition to classic bibliographic information such as authorship, title, year of publication, publisher, keywords, abstract, language, citations and references as well as information on access and usage rights, etc., PANGAEA datasets in schema.org are enriched with a range of additional information about the data to be described. These range from spatial coverage and temporal coverage to the description of parameters or variables (‘variableMeasured’), their units and underlying method information (‘measurementTechnique’), funding (‘funding’) and direct reference information (‘distribution’). Where possible, the corresponding entities are uniquely assigned with persistent identifiers such as ORCID IDs for individuals, ROR for institutions, DOI for references and samples (IGSN). Additionally, measurement variable descriptions are semantically defined with controlled terms of internationally recognized terminologies (‘hasDefinedTerm’).
The greatest potential for improving the description of PANGAEA datasets by schema.org or Bioschemas is seen in the adaptation of concepts for the structured description of the scientific sampling or observation event. The corresponding concept used by PANGAEA (‘event’) is already attributed to cultural events such as concerts in schema.org and therefore cannot be assigned accordingly. And at the time of writing this document, there are no other concepts available to fill this gap, neither in schema.org nor in Bioschemas.
Technically, PANGAEA’s own metadata schema is based on XML. To generate a schema.org representation, the in-house XML documents, which are stored in an Elasticsearch index, are transformed on-the-fly using an XSL transformation to an intermediate DOM tree (Document Object Model tree) which is serialized as JSON instead of XML. This PANGAEA-developed serialization component is serializing XML “elements” and “attributes” to JSON objects and arrays according to the following rules:
• Mixed type XML elements are transformed to JSON objects.
• Simple XML elements are transformed to JSON keys using the local name and strings as value, unless the XSL transformation emits an xsi:type attribute which is used as a hint for the correct data type.
• Repeated XML elements are merged during transformation and the values are converted to JSON arrays.
• XML attributes are converted to JSON keys using their local name prefixed with “@” and a string value.
• XML processing instructions emitted are used to insert arbitrary JSON structures using callbacks to the serialization code.
PANGAEA is planning to release the Java Code based on Java‘s DOM API, Jakarta XML Binding (JAXB) [44] and GSON [45] as Open Source software.
For compatibility reasons and due to the focus on earth and environmental data and the rather small proportion of biodiversity data, an enrichment of the schema.org representation of the data with elements from Bioschemas is currently not planned. Nonetheless, the aim is to maximize compliance with GFBio consensus elements in order to represent PANGAEA data in NFDI4Biodiversity and beyond in the best possible way.
The Staatliche Naturwissenschaftliche Sammlungen Bayerns (SNSB) is a GFBio Datacenter contributes more than 15 Million records to the NFDI4Biodiversity Network. The SNSB IT Center [46] develops and uses the Diversity Workbench, an integrated system for managing and publishing biodiversity data across various types. Data within the Diversity Workbench are stored in structured, relational database formats and organized as projects encompassing metadata for datasets. SNSB manages a variety of data types across all NFDI biodiversity categories (Types 1–5).
Type 1 data, collection specimens or units, are published using the ABCD standard. The datasets as a whole are published via ABCD archives. Single specimen records, expressed as ABCD:Units, are accessible through the BioCASe Provider Software with web pages providing the corresponding ABCD XML. However, these pages currently do not include Bioschemas markup. Notably, the stable identifiers issued via CETAF are exposed through the UnitGUID element within ABCD, enabling retrieval of RDF representations via content negotiation.
Type 2 data—taxonomic names and classifications—are managed within the Diversity Workbench’s taxonomy modules. These data are currently accessible as JSON via REST APIs and as Darwin Core Archives through GBIF channels, but neither ABCD nor Bioschemas serializations are presently provided.
For Type 3 data—environmental and ecological datasets—information structured within Diversity Workbench modules is published as SDD archives complemented by Ecological Metadata Language (EML) metadata. The landing pages for these datasets embed schema.org markup representing metadata derived dynamically from the underlying database. Moving forward, SNSB aims to enhance discoverability by incorporating Bioschemas representations both for ABCD archives and for unit-level records identified via CETAF stable identifiers.
This planned extension of Bioschemas markup will increase the interoperability and findability of SNSB’s biodiversity datasets across the broader research infrastructure.
The BEXIS2 data management system is a platform designed to store biodiversity data from projects or institutions involving several hundred users. It is a modular and extensible software that supports researchers throughout the data lifecycle, with a particular focus on data storage, dissemination, reuse, and discovery.12 BEXIS2 is a powerful tool for managing diverse types of biodiversity data, as it supports various data format types including tabular data, documents, GIS data, images, audio, and video files.
A total of 22 instances of the BEXIS2 data management platform are currently in use, primarily supporting the management and dissemination of environmental, biological, and ecological research data (Type 3). Within NFDI4Biodiversity, two of these instances are actively integrated as use cases. One is operated by the Bavarian Forest National Park [47] and hosts multi-taxon datasets, long-term climate records, and forest structure parameters. The second is part of the European Long-Term Ecosystem Research (eLTER) network, supporting the continuous collection and analysis of ecosystem processes. In addition, four further BEXIS2 instances are affiliated with NFDI4Biodiversity: the Amazon Tall Tower Observatory (ATTO) [48], the Kilimanjaro project, the University of Kassel (Department of Botany), and the Julius Kühn Institute (JKI).
The metadata schema for datasets is freely configurable for each instance and can be tailored to the specific requirements of different data formats and types. A basic set of metadata attributes such as data author, title and abstract are recommended, and several metadata schemas are provided by default (e.g. ABCD, DwC). The recommended basic set of metadata fields covers all minimum and some recommended properties of Bioschemas dataset profile (1.0-RELEASE).
To produce schema.org the appropriate metadata fields are mapped to schema.org attributes. The landing page of the datasets within BEXIS2 contains the metadata as harvestable schema.org. When creating the landing page of a dataset, the information is integrated in the page as JSON-LD.
After an initial test phase, the available set of properties will be expanded to include those that are commonly used across most BEXIS2 instances. This set can be further extended with additional attributes as needed. The mapping of metadata to Bioschemas must be carried out individually by each BEXIS2 instance and separately for each metadata schema in use within that instance.
In BEXIS2, metadata information of a dataset — either individual fields or the complete metadata set — can be mapped to different schemas, one of which is Bioschemas. The mapping is configured by an administrator and is currently based on the Bioschemas Dataset Profile (version 1.0-RELEASE). The core mapping includes all minimum and some recommended properties defined in the profile.
The mapping is extensible, and additional Bioschemas attributes may be integrated in the future.
The technical implementation strategy for mapping ABCD-compliant biodiversity data to Bioschemas within NFDI4Biodiversity is guided by a dual objective: establishing robust semantic interoperability for consortium-wide data discovery while supporting richer, cross-disciplinary reuse and integrability. This approach directly addresses requirements identified throughout the consortium, particularly the need to harmonize core metadata using the defined GFBio consensus elements (see also 2.4) as a minimum baseline for Type 1 data, and to enable variable levels of mapping detail for scenarios ranging from internal search and aggregation (“community case”) to comprehensive global data publication and reusability (“world case”). Recognizing that different discovery and application contexts necessitate tailored completeness of Bioschemas representations, the strategy incorporates both aggregation and contextual enrichment of ABCD elements, balancing search efficiency with information richness. By leveraging established infrastructure—especially the Data Transformation Service (DTS)—and promoting open documentation and sustained community engagement, the consortium aims to reduce implementation burdens, support transparency, and foster extensible technical solutions that will underpin current and future semantic alignment across NFDI4Biodiversity data flows.
A range of requirements have been identified concerning the mapping and transformation of ABCD-compliant biodiversity data to Bioschemas with respect to these two objectives and use cases. These requirements have been organized into functional and non-functional categories, each essential for laying a robust foundation for the technical integration of Bioschemas across NFDI4Biodiversity. The following paragraphs summarize these central requirements, accompanied by concise explanations.
1. Functional requirements
○ Type 1 data: Compliance with GFBio consensus elements
The mapping of ABCD GFBio consensus elements to Bioschemas is mandatory for a successful Bioschemas implementation, and a key requirement for the “community case”.
○ Varying detail grades
The two use cases require differing grades of detail concerning the mapping. For example, dataset search and indexing only require a defined and eventually summarizing (e.g. concerning taxa and spatial distribution) subset of both dataset- and unit-level metadata elements being mapped, whereas providing data and landing pages at dataset and unit-level for general discovery and reuse (the “world case”) require a more detailed or complete mapping.
○ Integration of missing types and properties in Bioschemas
For those elements where relevant ABCD concepts and properties do not have appropriate counterparts in Bioschemas, NFDI4Biodiversity will have to engage in Bioschemas community activities to propose and collaboratively refine them for inclusion in the Bioschemas portfolio. Until then, some of these elements could potentially be described differently (in the sense of provisionally circumscribed) or will have to be omitted with appropriate documentation for now.
○ Size of the Bioschemas representations
In order to ensure high performance when working with the Bioschemas representations, for example harvesting datasets for search, the mapping should ideally allow an aggregation of values for certain properties, for example, coordinates of ABCD Units into bounding boxes or taxon levels into groups of a higher rank. This ensures that the sizes of the mapping results are limited and manageable.
2. Non-functional requirements
○ Reducing implementation burdens
NFDI4Biodiversity aims to minimize the payload of a consortium-wide implementation of Bioschemas in terms of efforts for coordination, customisation and support by relying on established and operational infrastructure components such as the Data Transformation Service. The DTS already offers a range of transformations based on mappings between ABCD and CDM Light to PANGAEA PanSimple or ABCD (archive) to DwC Archive. And was extended recently to support Bioschemas-related transformations.
○ Documentation and public accessibility
To guarantee transparency, integrability, and accessibility of the primary interface provided by the DTS (API), the provision of an OpenAPI [49]-compliant documentation is mandatory.
○ Retention of transformations for asynchronous operations
Certain application scenarios for the DTS-based transformation pipeline, such as non-realtime uptake of Bioschemas-compliant mapping output, require temporary storage and provision of transformation results. In its current state, the DTS provides Bioschemas representations (in JSON-LD serialization) for a duration of 30 days after its creation.
This section provides a detailed exploration of the mapping process that translates ABCD version 2.06 compliant biodiversity datasets into Bioschemas representations, highlighting the approach’s scope, challenges, and current status.
Overview of mandatory, recommended and missing metadata elements
The mapping approach is anchored in the marginality concept defined by the Bioschemas profiles, distinguishing which properties are mandatory, recommended, or optional. These specifications are comprehensively documented in25 (Table 1 and Table 2), where each Bioschemas element is aligned with the corresponding ABCD element via its formal XML path [50]. This distinction reflects the hierarchical structure of the ABCD schema, where elements may pertain either to the dataset as a whole or to individual units.
An in-depth analysis demonstrates that all mandatory Bioschemas elements relevant to the Dataset profile can be effectively mapped from ABCD, ensuring compliance with essential data quality standards. Nevertheless, some recommended properties currently lack reliable counterparts in ABCD, indicating areas where further refinement or the introduction of new mappings might be necessary to achieve full semantic coverage.
Additionally, Table 3 of our comprehensive mapping25 summarizes ABCD elements without direct equivalents in Bioschemas. Many of these relate to metadata such as copyright statements, disclaimers, or detailed taxonomic name components, which are typically less critical for indexing but may be vital for comprehensive dataset landing pages or expanded metadata disclosures.
Transformation process: ABCD to Bioschemas
The core transformation pipeline uses a careful mapping strategy that balances semantic completeness with processing efficiency, with a primary focus on meeting the needs of search and indexing functionalities. The complete mapping rules outline how ABCD elements are linked to mandatory, recommended, and select optional Bioschemas properties that further enhance dataset discoverability.24
For instance, although the Bioschemas property spatialCoverage is classified as optional, its inclusion substantially improves search by enabling the use of geographic filters down to the country or region. Similarly, mapping elements such as the Identifier (Unit GUID) accommodates both URLs and plain text, employing the schema: URL or schema: Text types to ensure syntactic accuracy.
The transformation supports both straightforward one-to-one correspondences and complex mappings, which may involve aggregating several ABCD elements or expressing hierarchies via chained property paths indicated by colon separators in this manuscripts extended data tables.24 This approach preserves the essential semantic structure of the data while producing output files that are both comprehensive and manageable in size.
Beyond the primary focus on transforming biodiversity occurrence data (Type 1) from ABCD, the consortium recognizes the need to map a broader spectrum of biodiversity and environmental data types to corresponding Bioschemas and schema.org types. This is essential to enable comprehensive interoperability and to support diverse use cases across the NFDI4 Biodiversity community and beyond.
Table 2 summarizes the intended mapping targets for the major data types handled within the consortium. It also indicates the current progress status of these mappings.
For example:
• The BioSample type aligns well with ABCD units that describe physical specimens (preserved, living, fossil, or others).
• The Observation type can represent ABCD units describing observations, including those derived from human or machine detection.
• Taxon and TaxonName types are suited to describe taxonomic concepts and nomenclatural data, supporting rich semantic annotation of classifications and synonymy.
• LabProtocol addresses experimental and preparation procedures relevant, for instance, to microbial cultivation.
• PropertyValue caters to detailed trait measurements and attributes typically found in ecological or molecular datasets.
The consortium follows established best practices and existing guidelines, such as those proposed by the science-on-schema.org initiative [51], to ensure that mappings adhere to community standards and promote interoperability. Where metadata requires aggregation (e.g., summarizing multiple measurement values), recommendations suggest utilizing linked annotations such as schema: DataDownload to reference external detailed content.
Mapping completeness and specificity vary across types: mappings for Type 1 datasets are advanced, whereas those for other types, like media objects or comprehensive molecular metadata, are still under development or planned for future implementation. As an overarching principle, all mappings aim to comply with Bioschemas profile minimum requirements [52] to secure validation and broad acceptance within the semantic web ecosystem.
This multi-type mapping strategy positions Bioschemas as a flexible and extensible framework, capable of representing the full diversity of data managed within NFDI4Biodiversity and supporting the consortium’s vision of FAIR and interoperable biodiversity data infrastructures.
While the adoption of schema.org and Bioschemas brings significant benefits to semantic data description and interoperability within the consortium, several challenges and limitations have been identified that may affect implementation and use.
Technical challenges
Integrating the new Bioschemas transformation into existing services requires careful consideration of architectural constraints and workflows. It is essential to ensure that current transformation services remain unaffected during both development and deployment phases to avoid disruptions.
Performance is a critical factor, particularly when handling high volumes of data and real-time processing requirements. Although non-real-time data provision can rely on temporary storage, the system must strive for low latency transformation to support timely indexing and discovery.
Currently, the transformations use XSLT version 1.0, but upgrading to more recent versions (e.g., XSLT 2.0 or 3.0) can simplify the mapping logic and enable more robust processing features. However, this upgrade raises concerns about service versioning, compatibility, and the scope of necessary modifications within the existing transformation workflows.
Mapping challenges
Mapping between ABCD and Bioschemas involves complex considerations and careful bidirectional review, especially in cases where equivalent constructs are missing in Bioschemas. It is crucial that the mapping preserves as much information as possible to avoid loss of data fidelity.
Some ABCD elements currently lack direct counterparts in the Bioschemas vocabulary. The consortium plans to engage actively with the Bioschemas community to propose necessary new types or properties. Where immediate inclusion is not feasible, provisional approaches—such as using the additionalProperty attribute—can serve as workarounds, embedding unmapped attributes with detailed contextual information (e.g., including the original ABCD property name via the propertyID). The current progress of all mapping activities can be tracked and commented on via a dedicated GiHub repository [53].
To maintain reversibility of mappings, particularly for detailed use cases (beyond indexing-only scenarios), it is important to retain contextual cues that disambiguate merged or aggregated data. For example, when multiple ABCD elements map onto a single Bioschemas property, annotations indicating the original source elements facilitate back-mapping and semantic precision.
A notable example is the handling of measured variables (MeasurementOrFacts) in ABCD. While the Bioschemas variableMeasured property provides a general container for such data, careful treatment is needed to differentiate measurement types (e.g., altitude versus temperature), handle unit consistency, and manage aggregated values (e.g., min-max ranges). For use cases requiring a comprehensive mapping of ABCD content, measurements and other ABCD elements are handled differently. As illustrated in Figure 2, the mapping of unit-level data from ABCD 2.06 to the Bioschemas BioSample type avoids summarization of measurements; instead, diverse measurements are mapped to specific Bioschemas properties. For example, measurements related to location are mapped using the property locationCreated, while those concerning the species themselves are linked via the inverse property observationAbout.
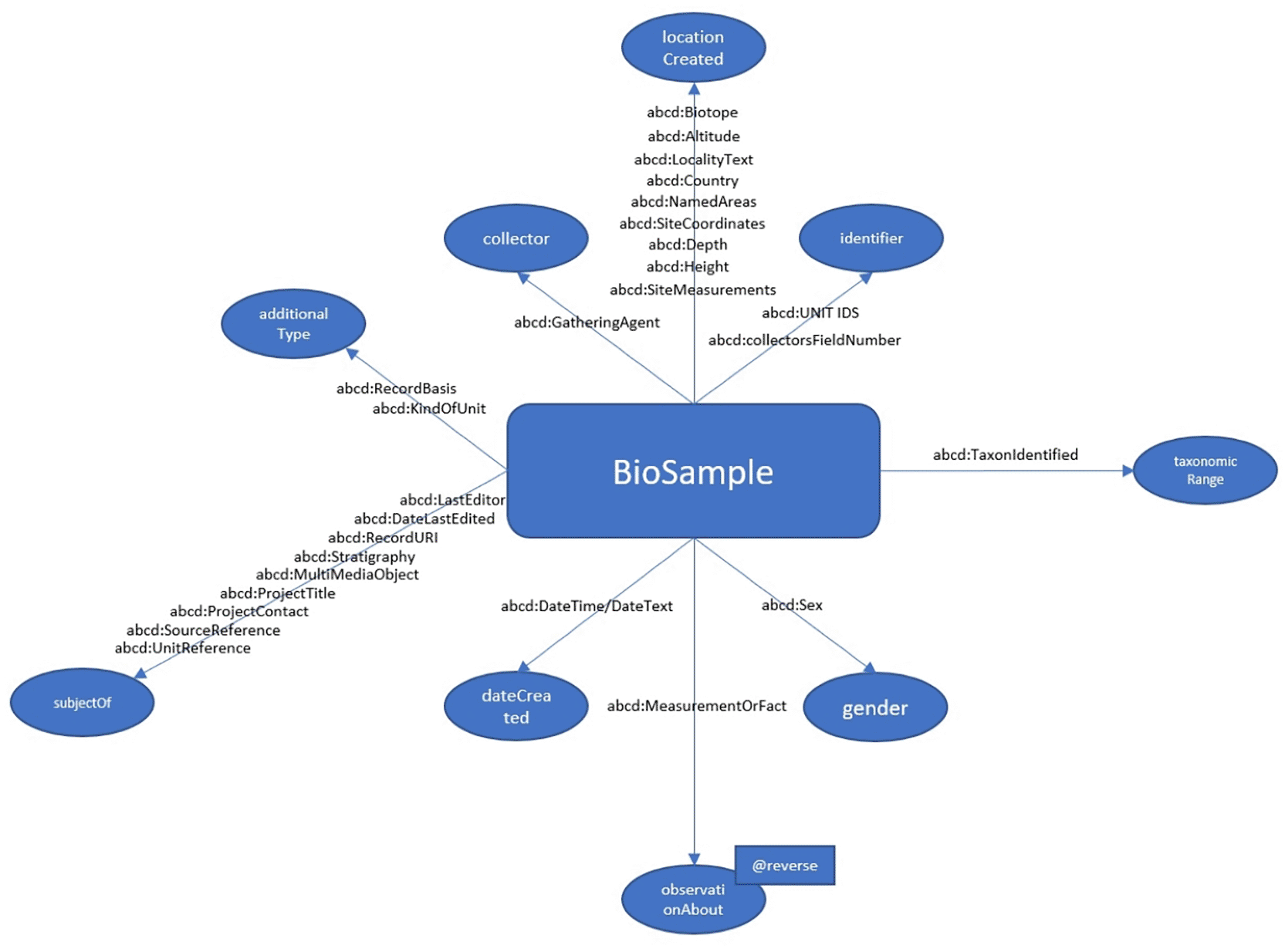
The current proposal for mapping ABCD:Unit elements to Bioschemas:BioSample properties preserving as much ABCD Unit content as possible during the transformation.
Several gaps remain where Bioschemas currently lacks suitable properties to fully capture certain ABCD elements25 (Table 3). Addressing these challenges requires active participation in the Bioschemas community to propose new types and properties, collaboratively refine their definitions, and advocate for their formal adoption within relevant standardization initiatives.
Limitations of current Bioschemas specifications
Certain essential biodiversity concepts—such as detailed observation events or specimen-specific properties—are not yet represented in Bioschemas. This gap complicates full semantic coverage and requires ongoing community engagement to extend the Bioschemas vocabulary.
The consortium has identified these gaps and is collaborating with Bioschemas developers to address them, including proposing new types and properties where necessary. Until these enhancements are adopted, some information will remain unmapped or must be encoded via generic extensibility mechanisms.
Future considerations
Balancing comprehensive data representation with practical constraints such as response size and processing time remains a challenge. Aggregation strategies, such as summarizing occurrences or clustering taxa, help manage data volume but need consistent standards.
The consortium recognizes that incremental improvements and community-driven refinements will be pivotal to realizing the full potential of Bioschemas-based interoperability within biodiversity research infrastructures.
The implementation of Bioschemas within the NFDI4Biodiversity consortium marks a significant step toward enhancing the discoverability and interoperability of biodiversity data for a broad spectrum of stakeholders, including the public, research institutions, industry, administrative bodies, and decision-makers. By semantically enriching metadata, we strive to ensure that resource discovery and evaluation—by both humans and machines—becomes more efficient, transparent, and inclusive. This approach aligns directly with NFDI4Biodiversity’s core mission to foster open access and maximize the utility of biodiversity data across diverse domains.
A central motivation for this work is to facilitate enhanced exchange of unified metadata within the consortium, while at the same time building the foundations necessary for interoperability with other domain-specific consortia, national and international initiatives. Strengthening such interoperability fosters a seamless interdisciplinary flow of information and helps to anchor the consortium within the wider global research ecosystem. Additionally, the initiative provides mechanisms to include valuable datasets that previously existed outside of major data centers, connecting these resources to the consortium’s search infrastructure and broader discovery mechanisms, such as Google Dataset Search. As a result, NFDI4Biodiversity moves toward a more complete and accessible representation of biodiversity data in Germany and beyond.
Technically, the consortium has focused on mapping the widely adopted ABCD metadata standard to Bioschemas, with the development of an automated transformation pipeline that generates Bioschemas-conformant representations from existing ABCD metadata. This automated process, aligned with GFBio consensus elements, not only minimizes the technical barriers for data providers but also ensures consistency, accuracy, and sustainability of semantic markups across the infrastructural landscape. Bioschemas metadata now underpins several critical infrastructure components, including data harvesting, the central search stack, the structured description of RDC data products (Aruna), and the construction and integration of knowledge graphs, thus strengthening both the internal infrastructure and its external interfaces.
Looking ahead, the consortium is committed to delivering an easy-to-use transformation pipeline service that enables data providers to efficiently generate Bioschemas representations ready for seamless integration on dataset landing pages using modern web technologies. A comprehensive implementation guideline will be provided, accommodating a range of user expertise and technical environments to foster inclusive adoption across all consortium members and partners. The consortium also aims to address remaining conceptual gaps in the semantic description of biodiversity data, for instance, regarding observation context and sampling events, through collaborative engagement with other NFDI consortia and the international Bioschemas and schema.org communities.
Moreover, ongoing efforts will focus on developing and formalizing formats and mechanisms for semantic and syntactic interoperability with related consortia. By institutionalizing collaboration and opening participation to interested parties from other communities, NFDI4Biodiversity aims to continue driving the harmonization and effective exchange of biodiversity information within and beyond the German National Research Data Infrastructure.
Through these actions, NFDI4Biodiversity positions itself as a forward-looking and collaborative force, dedicated to the advancement of open, interoperable, and FAIR biodiversity data. These efforts will support not just scientific research, but also evidence-based decision-making and societal engagement, catalyzing future developments in biodiversity informatics and ensuring that valuable ecosystem knowledge becomes increasingly accessible, actionable, and impactful.
AI-powered large language models (Perplexity.ai’s model Sonar based on Meta Llama 3.3 70B, Anthropic’s Claude Sonnet 4, and ChatGPT4 by OpenAI) were used to enhance language clarity and text flow in the writing process, supporting readability of the manuscript. All content has been reviewed and edited by the authors, who take full responsibility for the final manuscript.
Zenodo. Extended data for F1000 publication “Advancing FAIR Biodiversity Data: Bioschemas Implementation in NFDI4Biodiversity”. https://doi.org/10.5281/zenodo.17225215
The supplementary manuscript contains the following extended tables:
Table 1: Mandatory Bioschemas Dataset Elements with ABCD Property Equivalents
This table lists the mandatory properties required by the Bioschemas Dataset profile and shows, for each property, the corresponding source element(s) from the ABCD 2.06 metadata standard as used within NFDI4Biodiversity. It outlines the direct mapping paths supporting compliance with minimum Bioschemas requirements.
Table 2: Recommended Bioschemas Dataset Elements with ABCD Property Equivalents
This table presents the recommended (but non-mandatory) properties of the Bioschemas Dataset profile and details their mapping to ABCD elements, where applicable. It clarifies which elements can be populated from ABCD-compliant biodiversity datasets and highlights cases where no direct mapping exists.
Table 3: ABCD Dataset Elements Lacking Bioschemas Counterparts
This table identifies metadata fields present in the ABCD 2.06 standard that currently lack suitable or direct equivalents in the Bioschemas Dataset profile. It specifies which elements are missing in the semantic mapping and notes their potential relevance for search, indexing, or extended metadata publication scenarios.
Table 4: Detailed ABCD–Bioschemas Property Mapping for Dataset type
This table provides a comprehensive overview of how individual ABCD 2.06 elements are mapped to properties in the Bioschemas Dataset profile for indexing and harvesting purposes. Each row specifies the original ABCD element, the corresponding Bioschemas property, and the data type or mapping method applied, ensuring clarity and reproducibility of the transformation.
The respective publication is available under the terms of the Creative Commons Attribution 4.0 International license.
No supplementary materials, figures, datasets, or tables are associated with this publication.
No research data can be provided for this document because it is a conceptual and technical white paper describing implementation approaches, infrastructure integration, and metadata mappings, rather than presenting results from empirical data collection or original scientific experiments. All content is based on analyses, technical documentation, and synthesis of existing standards and tools, without the generation or use of new research datasets.
We thank the communities of schema.org and Bioschemas.org for their pioneering work to leverage web markup technologies to support the discoverability and FAIRness of scientific data and open knowledge transfer. We are grateful to Anton Güntsch and Roman Baum for their reviews, helpful feedback and comments.
[5] Extensible Markup language - a World Wide Web consortium (W3C) specification
[8] Resource Description Framework, https://www.w3.org/RDF
[9] JSON for linked data, https://json-ld.org/ - specified by the W3C JSON-LD Working Group
[10] Ecological Metadata Language,16 https://eml.ecoinformatics.org/
[11] Darwin Core Archive,15 https://dwc.tdwg.org/text/#21-the-archive-element
[12] Structured Descriptive Data - https://sdd.tdwg.org/
[25] https://transformation.gfbio.org/, also available on Github
[30] Open Archives Initiative Protocol for Metadata Harvesting, https://www.openarchives.org/pmh/
[32] A kind of extended Dublin Core, see also the PanSimple XML schema definition
[33] Such as https://ena2pansimple.gfbio.org/
[36] Common Data Model, https://cybertaxonomy.org/cdmlib/index.html#The_Common_Data_Model_-_CDM
[37] Description Language for Taxonomy, https://www.tdwg.org/standards/delta/
[38] DataCite Metadata Schema, https://schema.datacite.org/
[39] Minimum Information about any (x) Sequence,17 https://genomicsstandardsconsortium.github.io/mixs/
[42] FAIR Signposting Profile, https://signposting.org/FAIR/
[43] HTTP Content Negotiation, Mozilla Developer Network, https://developer.mozilla.org/en-US/docs/Web/HTTP/Guides/Content_negotiation
[44] Jakarta XML Binding v3.0, https://jakarta.ee/specifications/xml-binding/3.0/
[45] GSON, or Google Gson, is an open-source Java library that serializes Java objects to JSON, https://en.wikipedia.org/wiki/Gson
[46] https://snsb.info/
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the topic of the review discussed comprehensively in the context of the current literature?
Yes
Are all factual statements correct and adequately supported by citations?
Partly
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Knowledge engineering, FAIR data, Bioschemas
Is the topic of the review discussed comprehensively in the context of the current literature?
Yes
Are all factual statements correct and adequately supported by citations?
Yes
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Biodiversity data, Biodiversity Informatics, FAIR Implementation
Is the topic of the review discussed comprehensively in the context of the current literature?
Yes
Are all factual statements correct and adequately supported by citations?
Yes
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Biodiversity Informatics, Standards Development
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 27 Oct 25 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)