Keywords
nicotinamide mononucleotide, nicotinamide phosphoribosyltransferase, Vibrio natriegens, metabolic engineering
Nicotinamide mononucleotide (NMN), is a promising nutraceutical attracting much attention for its pharmacological and anti-aging efficacies. However, NMN-containing commercial products are very high-priced due to the lack of efficient and facile methods for industrial-scale production. To date, various metabolic engineering strategies have been successfully applied to produce NMN in Escherichia coli. Recently, Vibrio natriegens has become a promising host in the bioindustry thanks to its rapid growth and capabilities of broad substrate utilization. This study aims to evaluate the NMN biosynthesis capability of V. natriegens.
Firstly, a mutant V. natriegens strain (Δdns::araC-T7RNAP-KanR ΔpncC::FtnadE-SmR ΔnadR) was generated via multiplex genome editing by natural transformation (MuGENT). Nampt genes encoding nicotinamide phosphoribosyltransferase from Chitinophaga pinensis, Sphingopyxis sp. C-1, Haemophilus ducreyi, and Vibrio phage KVP40 were codon-optimized and cloned into pACYCDuet™-1 under the control of T7 promoter. The recombinant plasmids were electroporated into the mutant strain. The expression of recombinant NAMPTs in V. natriegens was evaluated by SDS-PAGE analysis and the intracellular NMN concentrations were quantified by HPLC.
After two rounds of MuGENT, V. natriegens V54-33 strain (Δdns::araC-T7RNAP-KanR ΔpncC::FtnadE-SmR ΔnadR) was successfully generated. SDS-PAGE analysis demonstrated that all NAMPTs were strongly expressed in the V54-33 strain. HPLC analysis revealed that the highest intracellular NMN concentration was obtained with NAMPT from Chitinophaga pinensis (44.5 μM), followed by NAMPT from Vibrio phage KVP40 (23.3 μM).
This study demonstrated the feasibility of NMN biosynthesis in V. natriegens.
nicotinamide mononucleotide, nicotinamide phosphoribosyltransferase, Vibrio natriegens, metabolic engineering
β-nicotinamide mononucleotide (NMN) is a precursor of NAD+, an essential coenzyme in various metabolic processes. NAD+ is involved in intracellular signaling pathways, regulating mitochondrial functions and other biochemical processes.1 In living organisms, intracellular NAD+ levels are directly influenced by its precursor, NMN. Recent studies reported various potential medical applications of NMN, particularly in anti-aging, cognitive-enhancing, and treatment of severe chronic diseases such as Alzheimer’s and diabetes.2,3
There are three major ways for NMN production: chemical synthesis, enzymatic methods and fermentation methods using genetically modified microbial strains.4 The chemical synthesis strategies offer the advantage of low production cost; however, the resulting isomeric compounds may exhibit toxicity or lack of biological activity. The enzymatic methods enhance selectivity and reduce toxicity, but the high cost of enzymes and phosphate sources such as ATP remain a major obstacle. Recent studies indicated that the fermentation approach using metabolically engineered microbial strains represents a promising solution to overcome the limitations of the latter methods. The most effective strategy for NMN biosynthesis in microbial cells primarily relies on: nicotinamide (NAM) supplement as a substrate, activation of pentose phosphate pathway to generate the precursor PRPP from carbon sources, overexpression of nicotinamide phosphoribosyltransferase (NAMPT) to catalyze the reaction between NAM and PRPP to produce NMN, and the expression of NMN transporter.5,6 Among these factors, NAMPT is the key enzyme that dictates NMN production efficiency.5–7 In a pioneer study, Marinescu et al. evaluated NAMPT from various sources and found that NAMPT from Haemophilus ducreyi, when expressed in E. coli BL21(DE3) pLysS, gave the highest intracellular NMN concentration (15.42 mg/L).7 Subsequently, Black et al. has combined the overexpression of both NAMPT and NMN synthase from Francisella tularensis (FtNadE) with the deletion of pncC and nadR, two genes involved in the catabolism of NMN, in E. coli, and achieved an intracellular NMN concentration of 1.5 mM.8 In another study, NAMPTs from ten organisms have been tested for the activity when expressed in E. coli and the results revealed that NAMPTs from Chitinophaga pinensis and Sphingopyxis sp. C-1 displayed significantly higher activities compared to NAMPTs derived from other sources.5 Similarly, Huang et al. performed a screening of eight NAMPTs from various sources and found that NAMPT from Vibrio phage KVP40 could biosynthesize 81.3 mg/L NMN intracellularly.6
Recently, Vibrio natriegens has emerged as a host for industrial biotechnology due to its extremely rapid growth rate with a reported doubling time of less than 10 min, non-pathogenic nature, genetic tractability, and ability to utilize a variety of carbon sources.9 In numerous studies, V. natriegens was used as an alternative expression system for recombinant protein production,10 and high-value metabolites such as pyruvate,11 2,3-butanediol.12 However, the application of V. natriegens in NMN production remains unexplored. Therefore, the present study aimed to evaluate the feasibility of NMN biosynthesis in V. natriegens.
Vibrio natriegens strain TND1964, kindly offered by Prof. Ankur Dalia from Indiana University, Bloomington, USA and Escherichia coli DH5α (Thermo Scientific™, cat. number EC0112) were used as hosts for expression and cloning, respectively.
Oligonucleotides were synthesized by Macrogen (Korea). Expression vector used in this study was pACYCDuet™-1 (Novagen, cat. number 71147-3).
All other reagents were obtained from Thermo Scientific, New England Biolabs, Merck, Qiagen, and Bio Basic, unless otherwise specified.
Vibrio natriegens V54-33 strain (Δdns::araC-T7RNAP-KanR ΔpncC::FtnadE-SmR ΔnadR) was generated using multiplex genome editing by natural transformation (MuGENT) method.13 Details were presented in the Supplement I (refer to extended data).14
The Nampt genes encoding nicotinamide phosphoribosyltransferase (NAMPT) from C. pinensis, Sphingopyxis sp. C-1, H. ducreyi, and Vibrio phage KVP40, were codon-optimized for expression in V. natriegens and synthesized by Genscript. These genes were cloned into the pACYCDuet™-1 ( Novagen, cat. number 71147 ) under the control of T7 promoter-1 (extended data- Supplement II).14
The preparation of V. natriegens electrocompetent cells and the electroporation of DNA plasmids were carried out following the procedures described by Weinstock et al.15 Briefly, a V54-33 colony was grown in LBv2 medium at 30°C, 200 rpm overnight. LBv2 is LB-Miller (Bio Basic, cat. number SD7003) supplemented with v2 salts (204 mM NaCl (Bio Basic, cat. number DB0483), 4.2 mM KCl (Bio Basic, cat. number PB0440, and 23.14 mM MgCl2 (Bio Basic, cat. number MB0328)). Subsequently, 1% (v/v) of the overnight culture was reinoculated into 50 mL LBv2 fresh medium and incubated at 30°C, 200 rpm until OD600 reached 0.5. The culture were centrifuged at 6500 rpm for 20 min at 4°C and cells were washed three times with 1 mL of electroporation buffer (680 mM sucrose (Bio Basic, cat. SB0498), 7 mM K2HPO4 (Bio Basic, cat. PRB0447), pH 7). Cells were aliquoted (100 μL per vial), frozen in dry ice, and stored at −80°C. For transformation, a vial of electrocompetent cells is thawed on ice and mixed with 50 ng of plasmid DNA. The mixture is transferred to a 0.1 cm gap electroporation cuvette and pulsed at 700 V, 25 μF, 200 Ω. After electroporation, cells are immediately resuspended in 500 μL recovery medium (LBv2, 680 mM sucrose) and incubated at 30°C for 1 h. Transformants were selected on LBv2 agar plates supplemented with Chloramphenicol (25 μg/mL) (Bio Basic, cat. number CB0118).
A single colony was inoculated into 2 mL of LBv2 medium supplemented with Chloramphenicol (25 μg/mL) (Bio Basic, cat. number CB0118) and cultured at 30 °C, 200 rpm overnight. Subsequently, 1% (v/v) of the overnight culture was reinoculated into 10 mL of fresh LBv2 at 30 °C and 200 rpm. Induction was carried out by adding 2 g/L arabinose (Bio Basic, cat. number AB0071L) when OD600 reached 0.6−0.8. The cultures were cultivated until OD600 reached 5-6 and the cultured cells, corresponding to 5 mL cultures, were collected for SDS-PAGE analysis. The remaining cultures were centrifuged at 3000 g for 5 min at 4 oC. After discarding the supernatant, the cells were washed in 20 mL modified M9 medium16 and resuspended in 5 mL modified M9 medium supplemented with Chloramphenicol (25 μg/mL) (Bio Basic, cat. number CB0118), 1 mM nicotinamide (Sigma, cat. number N0636) and 1 mM nicotinic acid (Sigma, cat. number N0765). The cells were cultured at 30 oC and 200 rpm for 5 h and cultured cells, corresponding to 1 OD600, were collected, washed with 1 mL of water and stored at -80 oC until NMN quantification.
NMN quantification was carried out according to the method described by Vu et al.17 Briefly, cells were resuspended in 500 μL of water and lysed by sonication on ice by using VC 130 System (Sonics & Materials, Inc, cat. number VC130). The amplitude was set at 80% and on/off pulse of 10 s duration each was given for 1 min. Subsequently, 100 μL of trichloroacetic acid 25% (Bio Basic, cat. number TB0968) was added to inactivate the enzymes in the lysate. The obtained mixture was clarified by centrifugation at 15000 g for 15 min at 4 oC and the supernatant was used for HPLC analysis. Supernatants were run on a Shimadzu LC-20A high-performance liquid chromatography (HPLC) system with a ReliantTM C18 chromatography column (150 mm x 4.6 mm, 5 μm) (Waters, cat. number 186007282). Mobile phase used for separation was 10 mM phosphate buffer pH 3: methanol (90:10) (Supelco, cat. number 106007), at the flow rate of 1.0 mL/min. The injection volume was 20 μL, and individual peak areas were determined using a photo diode array at 261 nm.
In our present study, a metabolic engineering approach ( Figure 1) combining the insertion of key enzymes in the NMN biosynthetic pathway (NMN synthase from Francisella tularensis (FtNadE) and the most promising NAMPTs from previous studies) and the inactivation of endogenous NMN-degrading enzymes (NMN amidohydrolase, encoded by pncC, and NMN adenylyltransferase, encoded by nadR) was used to evaluate the NMN production capability in V. natriegens. To this end, a mutant strain, called V54-33, has been successfully generated by two rounds of MuGENT introducing araC-T7 RNA polymerase construct and FtNadE under the control of T7 promoter, respectively, while deleting pncC and nadR genes (extended data- Supplement I).14 This allowed a tight induction of NAMPTs and FtNadE by arabinose, and thereby provided a controlled regulation for NMN biosynthesis. Of note, our previous attempts using T7 IPTG-induced system were unsuccessful because of the growth inhibition in transformants (possibly due to the leaky expression of T7lac promoter) (data not shown).
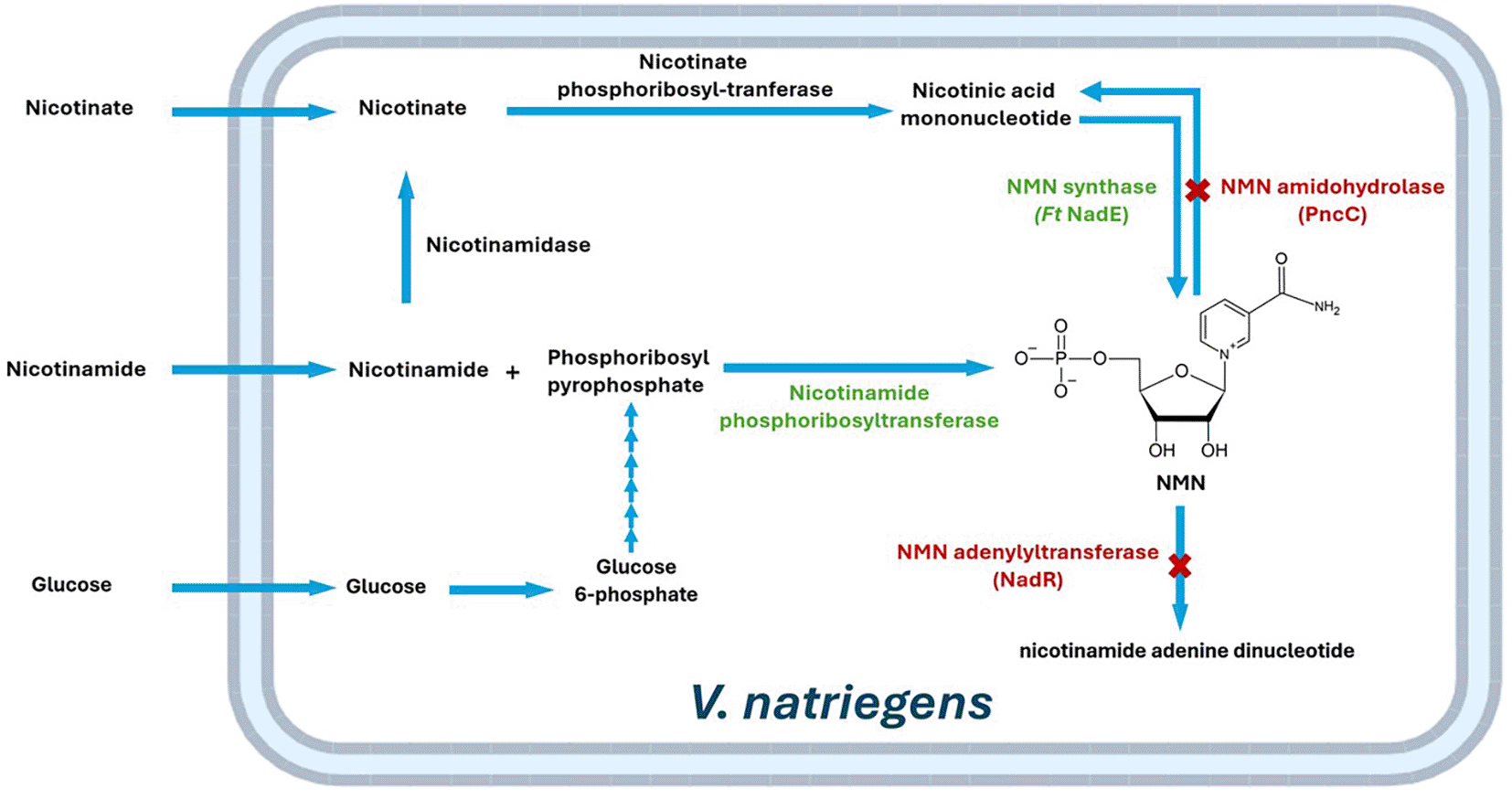
NMN: nicotinamide mononucleotide.
Subsequently, four plasmids overexpressing NAMPTs from C. pinensis (CP), Sphingopyxis sp. C-1 (SSC), H. ducreyi (HD) and Vibrio phage KVP40 (VP) under the control of T7 promoter, were successfully generated (extended data-Supplement II).14 These four enzymes have been reported as the most efficient NAMPTs in separated studies.5–7 In the next step, the strain V54-33 has been transformed with these plasmids harboring NAMPTs. SDS-PAGE analysis ( Figure 2) clearly showed that all recombinant strains carrying NAMPT plasmids displayed an intense band at the expected size of 56 kDa after induction with arabinose, while the original V54-33 strain did not possess this band. These results demonstrated that NAMPTs from CP, HD, SSC and VP were successfully expressed in V. natriegens.
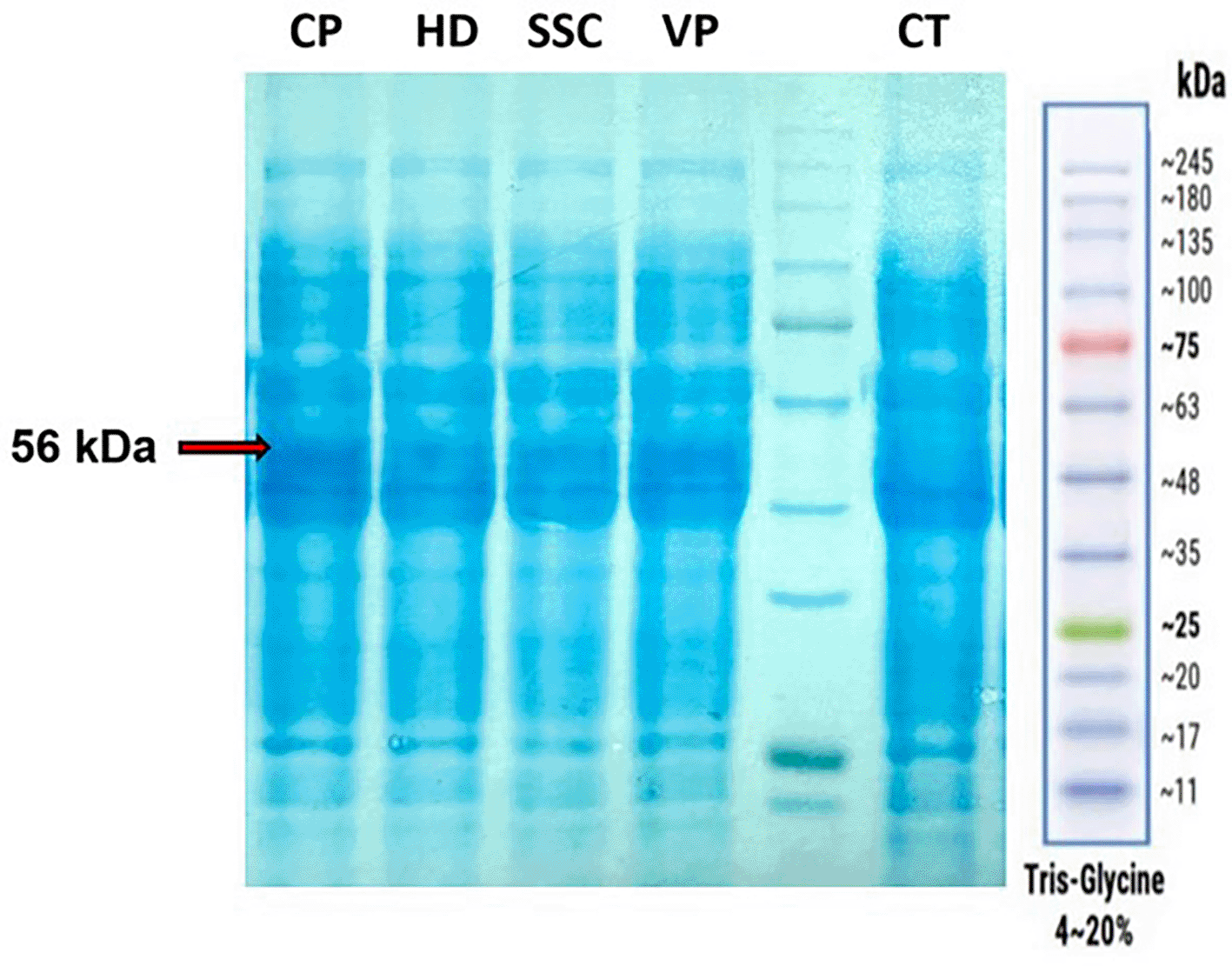
L: GangNam-STAIN™ Prestained Protein Ladder. CP: lysate of V54-33 strain overexpressing NAMPT from Chitinophaga pinensis. HD: lysate of V54-33 strain overexpressing NAMPT from Haemophilus ducreyi. SSC: lysate of V54-33 strain overexpressing NAMPT from Sphingopyxis sp. C-1. VP: lysate of V54-33 strain overexpressing NAMPT from Vibrio phage KVP40. CT: lysate of V54-33 strain.
These recombinant strains were then transferred into modified M9 medium to assess NMN biosynthesis. HPLC quantification revealed that three out of four transformants (CP, HD, and VP) were capable of intracellular NMN accumulation, while SSC transformants and the V54-33 strain exhibited NMN levels below the limit of detection (LOD) of the method ( Figure 3, extended data-Supplement III).14 Among them, CP transformants exhibited the highest intracellular NMN titers (44.5 μM), whereas HD transformants showed significantly lower NMN accumulation (12.1 μM, p = 0.0111) ( Figure 3, extended data- Supplement III).14 The NMN concentration of VP transformants was intermediate (23.3 μM) (extended data- Supplement III),14 which aligns with prior research by Huang et al., showing that NAMPT from VP was more efficient in NMN accumulation than NAMPT from HD.6 When considering the absolute NMN titers reported in E. coli, NMN accumulation in CP transformants was comparable to that reported by Marinescu et al. (15.42 mg/L),7 but significantly lower than those reported by Black et al. (1.5 mM)8 and Huang et al. (81.3 mg/L).6 These disparities could be explained by the difference in expression systems: (i) FtNadE was expressed on a multiple-copy plasmid in E. coli in the study by Black et al.8 while this gene was incorporated into the chromosome of V. natriegens as a single-copy in our study; (ii) ushA encoding 5′-phosphatase capable of dephosphorylation of NMN to nicotinamide riboside and purR regulating the synthesis of the precursor phosphoribosyl pyrophosphate (PRPP) were knocked out in the study of Huang et al.6 whereas these genes have not been inactivated in the present study.
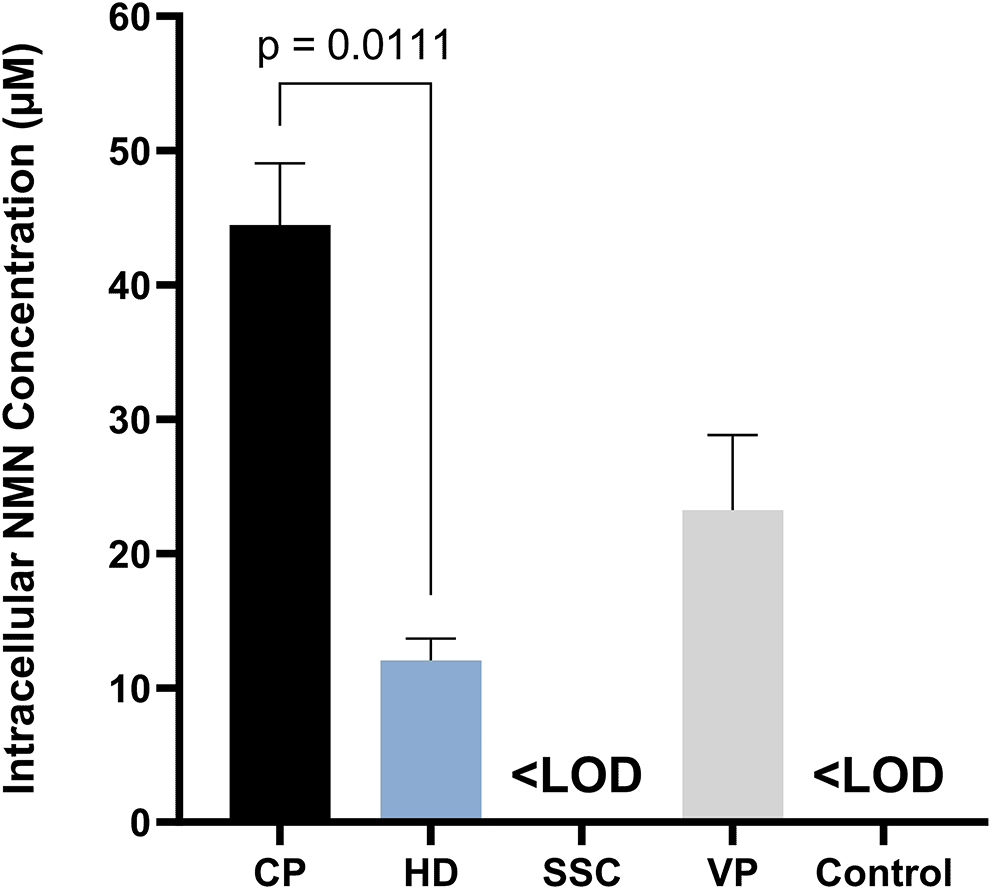
CP: V54-33 strain overexpressing NAMPT from Chitinophaga pinensis. HD: V54-33 strain overexpressing NAMPT from Haemophilus ducreyi. SSC: V54-33 strain overexpressing NAMPT from Sphingopyxis sp. C-1. VP: V54-33 strain overexpressing NAMPT from Vibrio phage KVP40. Control: V54-33 strain. <LOD: concentration lower than limit of detection of HPLC method. Data are expressed as mean ± S.D. (error bars). Statistical analysis of differences between groups was performed by unpaired t-test (p < 0.05).
In summary, this study demonstrated the feasibility of NMN biosynthesis in V. natriegens. Further research focusing on the activation of pentose phosphate pathway and the transporters of NMN and NAM is underway to improve NMN production in this promising host.
Dataset from NCBI Sequence Read Archive: Sequencing results for bioproject: “β-nicotinamide mononucleotide production in Vibrio natriegens: a preliminary study”.
Accession number PRJNA1231393; https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA1231393.
Figshare: Supplement for “β-nicotinamide mononucleotide production in Vibrio natriegens: a preliminary study”.14
Doi: https://doi.org/10.6084/m9.figshare.28547603.v1
This project contains the following extended data:
• Supplement I: Generation of Vibrio natriegens strain V54-33 (Δdns::araC-T7RNAP-KanR ΔpncC::FtnadE-SmR ΔnadR).pdf
• Supplement II: Generation of pACYCDuet-Nampt plasmids.pdf
• Supplement III: Quantification of intracellular NMN concentration by HPLC.pdf
Data are available under the terms of the CC BY 4.0
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: microbiology, bacterial and yeast metabolic engineering
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Biochemistry, Natural Product Chemistry, Cofactors
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Biotechnology, Microbiology, Cofactors
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 07 Apr 25 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)