Keywords
emerging; pathogen; acute respiratory infection; children; Vietnam
This article is included in the Pathogens gateway.
The COVID-19 pandemic has caused changes in respiratory infectious diseases. Examining the patterns of pathogens associated with acute respiratory infection (ARI) in children before, during, and after the COVID-19 pandemic would help to understand the impact of the pandemic on pathogen emergence or re-emergence.
We analyzed de-identified data from microbiology assays of nasopharyngeal and blood samples of children ≤15 years old with ARI who visited Vinmec Times City International Hospital in Hanoi, Vietnam from 01/01/2019 to 31/12/2023. The data were aggregated by month, and time-series analysis and visualization were performed.
A Bacterial Polymerase Chain Reaction (PCR) panel was performed on 4,125 samples (67% positive), Mycoplasma pneumonia (MP) IgM was performed on 5,049 samples (39% positive), bacterial culture was performed on 10,280 samples (43% positive), and viral PCR or rapid test was performed on 42,300 samples (23% positive). After the COVID-19 pandemic from mid-2022, Haemophilus influenzae (HI) and Streptococcus pneumoniae (SP) have re-emerged as epidemic pathogens associated with lower respiratory tract infection (LRI). Influenza type A and type B have re-established regular cycles of peaks in winter-spring months after an early rebound together with an unprecedented new emergence of Human Adenovirus (HAdV) soon after the relief of COVID-19 restriction in mid-2022. Late after the COVID-19 pandemic, from mid-2023, atypical pneumonia pathogen Mycoplasma pneumonia (MP) has emerged remarkably and has become epidemic; there was also a small, brief emergence of Chlamydophila pneumoniae (CP) infection.
Our data characterize the influence of the COVID-19 pandemic on the patterns of respiratory infection pathogens in children and is useful for disease surveillance and public health interventions.
emerging; pathogen; acute respiratory infection; children; Vietnam
The COVID-19 pandemic and related policies have changed the patterns of respiratory infectious diseases.1–3 The incidence of most respiratory infections, apart from COVID-19, has decreased during the COVID-19 pandemic due to restricted measures.2 After easing COVID-19 related restricted measures, the sudden increase of exposure in the community, whose immunity might be waned during the prolonged no-contact quarantine period, might lead to irregular respiratory infection patterns.1 There have been some reports regarding the rebound of viral pathogens, such as influenza,4,5 and bacterial pathogens, such as Mycoplasma,6 after the COVID-19 pandemic. We previously reported the molecular data of a case series of Human Adenovirus (HAdV) outbreak in 20227 and Mycoplasma pneumonia (MP) outbreak in 20238 in Hanoi, Vietnam.
Knowing how the COVID-19 pandemic and related restrictions influenced the pattern of viral and bacterial pathogens associated with acute respiratory infections (ARIs) in children would be helpful for public health interventions during the post-COVID-19 era. This could also provide useful information for health authorities to cope with future pandemics. Some studies have described the impact of COVID-19 on the pattern of pathogens associated with ARIs in children, including those in Italy9 and China.10–14 However, published data regarding this matter in Vietnam are still scarce. As Vietnam may be different from other countries in terms of the COVID-19 outbreak situation and related restricted measures as well as population characteristics, such studies specifically in Vietnam are necessary.
In this report, we describe the overall pattern of emerging viral and bacterial pathogens associated with ARIs in children before, during, and after the COVID-19 pandemic in Hanoi, Vietnam, to understand the influence of the COVID-19 pandemic and related restrictions on the emergence or re-emergence of other respiratory infection pathogens in children.
This study was conducted after obtaining ethical approval (approval No 110/2024/CN/HDDD VMEC, date October 10, 2024) from the Ethical Committee of Vinmec Healthcare System and VinUniversity (Ethical Committee establishment decision No 24/2016/QD-VINMEC). Informed consent was waived by the Ethical Committee for retrospective analysis and report of de-identified data.
We analyzed the deidentified data of microbiology assays of nasopharyngeal and blood samples of children with ARIs who visited Vinmec Times City International Hospital in Hanoi, Vietnam, from January 1, 2019, to December 31, 2023. This study data period included the time before, during, and after the COVID-19 pandemic, and thus facilitated the evaluation of the impact of the COVID-19 pandemic on the patterns of pathogens associated with ARIs in children. Data were obtained from the recorded microbiology assay result database of the Vinmec Microbiology Department. The original data contained information regarding patient identification, sex, date of birth, date of microbiological assays, diagnosis of microbiological assay indication, and assay results. The data were de-identified before being transferred to the research team, which had no access to information that could identify individual participants during data access and analysis. All patients included in the analysis were ≤15 years old with one or more diagnoses of respiratory infection and with one or more microbiological assays of nasopharyngeal or blood samples. All included patients were from Hanoi and surrounding provinces.
During the study period in Hanoi, Vietnam, due to the COVID-19 pandemic, intermittent social distancing was applied from mid-2020 to the first half of 2021, when there were only sporadic COVID-19 cases or clusters, and there was no remarkable COVID-19 outbreak. The lockdown was then applied in the second half of 2021 due to the severe COVID-19 outbreak in Vietnam during this time.15 After the lockdown period, milder social distancing was applied in early 2022, when most of the population had been vaccinated with COVID-19 vaccines, and most of the cases were mild and Omicron was the dominant reported variant. Then COVID-19 related restriction was relieved starting from the second quarter of 2022, when COVID-19 was no longer a public health concern in Vietnam.16 The timeline associated with COVID-19 activity and COVID-19 related restricted measures is shown in Figure 1. Our study data included the periods before, during and after the COVID-19 related restriction and thus enabling us to examine the impact of the COVID-19 pandemic on the pattern of pathogens associated with ARIs in children.
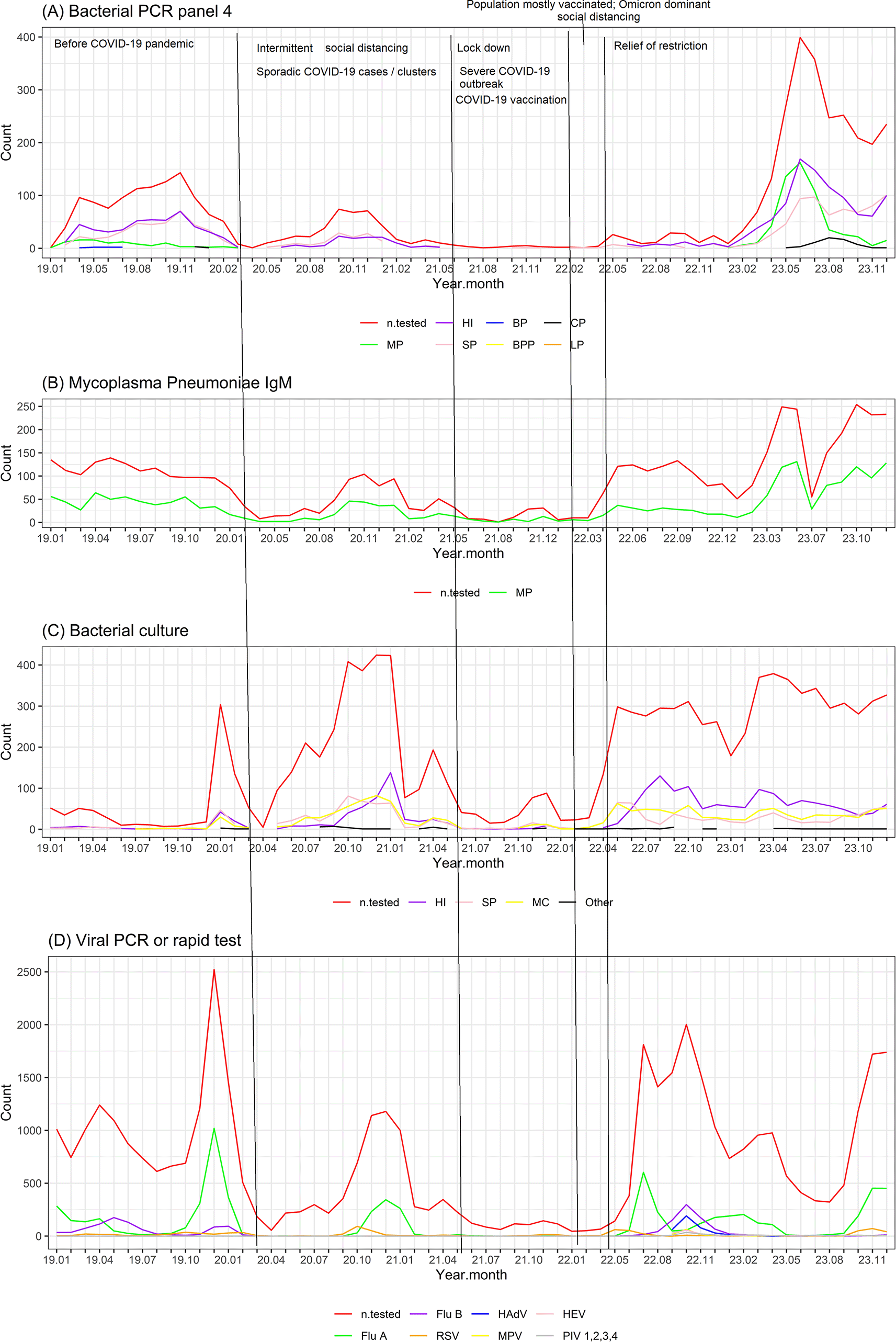
(A) Allplex Respiratory Panel 4 (https://www.seegene.com/assays/allplex_respiratory_panel_4 ) using multiplex one-step real-time Polymerase Chain Reaction (PCR) of nasopharyngeal samples for the detection of 7 bacteria causing respiratory tract infections including Bordetella parapertussis (BPP), Bordetella pertussis (BP), Chlamydophila pneumoniae (CP), Haemophilus influenzae (HI), Legionella pneumophila (LP), Mycoplasma pneumoniae (MP), Streptococcus pneumoniae (SP). The plot shows the number of patients tested (n.tested) and the number of patients positive for each pathogen.
(B) Blood IgM serology test for Mycoplasma pneumoniae (LIAISON Mycoplasma pneumoniae IgM of Diasorin, Ireland). The plot shows the number of patients tested (n.tested) and the number of patients with IgM seropositive for Mycoplasma pneumoniae.
(C) Bacterial culture of nasopharyngeal samples (following the procedures guided in “Clinical Microbiology Procedures Handbook” of American Society for Microbiology17). The plot show the number of patients tested (n.tested) and the number of patients with positive culture for each pathogen. Haemophilus influenzae (HI), Streptococcus pneumoniae (SP), Moraxella catarrhalis (MC) and other pathogens. Figure 1. Time series plots of pathogens associated with ARIs in children from 2019 to 2023. (A) Allplex Respiratory Panel 4 (https://www.seegene.com/assays/allplex_respiratory_panel_4 ) using multiplex one-step real-time Polymerase Chain Reaction (PCR) of nasopharyngeal samples for the detection of 7 bacteria causing respiratory tract infections including Bordetella parapertussis (BPP), Bordetella pertussis (BP), Chlamydophila pneumoniae (CP), Haemophilus influenzae (HI), Legionella pneumophila (LP), Mycoplasma pneumoniae (MP), Streptococcus pneumoniae (SP). The plot shows the number of patients tested (n.tested) and the number of patients positive for each pathogen. (B) Blood IgM serology test for Mycoplasma pneumoniae (LIAISON Mycoplasma pneumoniae IgM of Diasorin, Ireland). The plot shows the number of patients tested (n.tested) and the number of patients with IgM seropositive for Mycoplasma pneumoniae. (C) Bacterial culture of nasopharyngeal samples (following the procedures guided in “Clinical Microbiology Procedures Handbook” of American Society for Microbiology17). The plot show the number of patients tested (n.tested) and the number of patients with positive culture for each pathogen. Haemophilus influenzae (HI), Streptococcus pneumoniae (SP), Moraxella catarrhalis (MC) and other pathogens. (D) Viral assays including Allplex Respiratory PCR Panel 1 (https://www.seegene.com/assays/allplex_respiratory_panel_1 ) using multiplex one-step real-time RT-PCR for the detection of Influenza type A (Flu A), Influenza type B (Flu B) and Respiratory Syncytial Virus (RSV), or rapid test for the detection of Flu A and Flu B (SD Bioline Influenza A/B Ag of Standard Diagnostic, South Korea), and RSV (BD Veritor System RSV Devices of Becton, Dickinson and Company, USA) or Allplex Respiratory PCR Panel 2 (https://www.seegene.com/assays/allplex_respiratory_panel_2 ) using multiplex one-step real-time RT-PCR of nasopharyngeal samples for the detection of 7 viruses causing respiratory tract infections including Human Adenovirus (HAdV), Enterovirus (HEV), Metapneumovirus (MPV), Parainfluenza virus 1,2,3 4 (PIV1,2,3,4). The plot shows the number of patients tested (n.tested) and the number of patients positive for each pathogen. The x-axis shows the year and month of the year (e.g. 19.01 means January 2019, 23.10 means October 2023). Co-detection: ≥2 pathogens were detected. ARIs: acute respiratory infections.
(D) Viral assays including Allplex Respiratory PCR Panel 1 (https://www.seegene.com/assays/allplex_respiratory_panel_1 ) using multiplex one-step real-time RT-PCR for the detection of Influenza type A (Flu A), Influenza type B (Flu B) and Respiratory Syncytial Virus (RSV), or rapid test for the detection of Flu A and Flu B (SD Bioline Influenza A/B Ag of Standard Diagnostic, South Korea), and RSV (BD Veritor System RSV Devices of Becton, Dickinson and Company, USA) or Allplex Respiratory PCR Panel 2 (https://www.seegene.com/assays/allplex_respiratory_panel_2 ) using multiplex one-step real-time RT-PCR of nasopharyngeal samples for the detection of 7 viruses causing respiratory tract infections including Human Adenovirus (HAdV), Enterovirus (HEV), Metapneumovirus (MPV), Parainfluenza virus 1,2,3 4 (PIV1,2,3,4). The plot shows the number of patients tested (n.tested) and the number of patients positive for each pathogen.
The x-axis shows the year and month of the year (e.g. 19.01 means January 2019, 23.10 means October 2023).
Co-detection: ≥2 pathogens were detected.
ARIs: acute respiratory infections.
The microbiology assays included in the analysis were:
(A) Allplex Respiratory Panel 4 (https://www.seegene.com/assays/allplex_respiratory_panel_4 ) using real-time Polymerase Chain Reaction (PCR) of nasopharyngeal samples to detect seven bacteria causing respiratory tract infections, including Bordetella parapertussis (BPP), Bordetella pertussis (BP), Chlamydophila pneumoniae (CP), Haemophilus influenzae (HI), Legionella pneumophila (LP), Mycoplasma pneumoniae (MP), Streptococcus pneumoniae (SP).
(B) Blood IgM serology test for Mycoplasma pneumonia (LIAISON Mycoplasma pneumoniae IgM; Diasorin, Ireland).
(C) Bacterial culture of nasopharyngeal samples (following the procedures guided in the “Clinical Microbiology Procedures Handbook” of the American Society for Microbiology17).
(D) Viral assays including Allplex Respiratory PCR Panel 1 (https://www.seegene.com/assays/allplex_respiratory_panel_1 ) using multiplex one-step real-time RT-PCR for the detection of Influenza type A (Flu A), Influenza type B (Flu B), and Respiratory Syncytial Virus (RSV), or rapid test for the detection of Flu A and Flu B (SD Bioline Influenza A/B Ag of Standard Diagnostic, South Korea), and RSV (BD Veritor System RSV Devices of Becton, Dickinson and Company, USA) or Allplex Respiratory PCR Panel 2 (https://www.seegene.com/assays/allplex_respiratory_panel_2 ) using multiplex one-step real-time RT-PCR of nasopharyngeal samples for the detection of seven viruses causing respiratory tract infections, including Human Adenovirus (HAdV), Enterovirus (HEV), Metapneumovirus (MPV), Parainfluenza virus 1,2,3 4 (PIV1,2,3,4).
Clinical data accompanying the microbiological assay data available for our analysis were patient diagnosis, age, and sex. Patients were classified into two groups: upper respiratory infections (URI) and lower respiratory infections (LRI) based on the diagnoses for indication of microbiology assays. LRI included those with the following diagnoses: pneumonia, bronchitis, bronchiolitis, bronchopneumonia, and asthma. Those with a diagnosis of upper respiratory tract infection and without any diagnosis involving the lower respiratory tract were classified as having URI. All participants included in the analysis must have data regarding the results of at least one microbiological assay using nasopharyngeal or blood samples, the date when the assays were performed, diagnosis of ARIs, age, and sex. Those with missing data for one or more variables were excluded from the analysis.
The data were aggregated by month, and time-series plots were used to visualize the data by month from January 2019 to December 2023. The number of samples tested and the number of samples positive for each pathogen were plotted by month. For patient characteristics, the mean age with standard error, percentage of males, and percentage of diagnosis category LRI were plotted. Co-detection (≥ 2 pathogens detected in a patient) was also described. Patient characteristics were compared between the groups using the Kruskal–Wallis test for continuous variables and the chi-square test for categorical variables.
In overall, in the beginning of the pandemic time from early 2020 to the first half of 2021, when intermittent social distancing was applied and when there were sporadic COVID-19 cases or clusters in Hanoi, Vietnam, the number of ARI cases positive for bacterial or other viral pathogens decreased as compared to that before the COVID-19 pandemic. However, there were still peaks of both bacterial and other viral pathogens from winter 2020 to spring 2021, although the peak of other viral pathogens was smaller than that before COVID-19. An exception was the relatively high number of bacterial cultures positive for SP, HI, and MI at the end of 2020 and early 2021. During the lockdown period in the second half of 2021 due to the severe COVID-19 outbreak in Vietnam, the reduction was much more remarkable in that the number of ARI cases positive for bacterial or other viral pathogens was almost close to zero. Both bacterial and other viral pathogens started to increase after the relief of COVID-19 related restriction starting from the second quarter of 2022. However, the rise of viral pathogens was sooner and faster, resulting in a high peak just a few months after the relief of COVID-19 related restriction ( Figure 1).
Bacterial PCR Panel 4 was performed on 4125 samples from 3708 patients, of which 2762 (67%) samples were positive for ≥1 pathogen tested in the panel. LRI accounted for 73%, males accounted for 58%, and the average age was 3.2 years. A total of 1606 (39%) samples were positive for HI, 1237 (30%) for SP, 675 (16%) for MP, and 777 (18.8%) for ≥2 pathogens (co-detection). Co-detection was observed in 44% of those positive for MP, 49% of those positive for SP, and 41% of those positive for HI. Nearly all (90%) of those positive for MP had LRI, and approximately three-fourths of those positive for SP or HI had LRI. The average age of patients positive for SP or HI was less than 3 years, while the average age of those positive for MP was nearly 5 years (p < 0.001) ( Table 1, Figure 2A). There was one peak of HI and SP infections at the end of 2019 before the COVID-19 pandemic and a smaller peak at the end of 2020 before a close-to-zero flat period during the severe COVID-19 outbreak in the second half of 2021 in Vietnam. After the relief of COVID-19 related restriction starting from the second quarter of 2022, a much higher peak of HI and SP infection re-emerged in the middle of 2023 in parallel with a newly emerged large peak of MP infection. A new small peak of CP infection also briefly emerged a few months later, in the middle of 2023 ( Figure 1A).
| Bacterial pathogens | Viral pathogens | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR Panel 4a | MP IgMb | Culturec | PCR or rapid testd | PCR Panel 2d | ||||||||||||||||||||||
| Patient characteristics | MP Positive (N = 675; 16%) | SP Positive (N = 1237; 30%) | HI positive (N = 1606; 39%) | Co-detection (N = 777; 19%) | Total panel 4 (N = 4125) | p value* | MP IgM Positive (N = 1976; 39%) | HI positive (N = 1878; 18%) | SP positive (N = 1230; 12%) | MC positive (N = 1367; 13%) | Co-detection (N = 177; 1.7%) | Total culture (N = 10280) | P value* | Patient characteristics | Flu A positive (N = 6614/40112; 17%) | Flu B positive (N = 1677/40112; 4%) | RSV positive (N = 810/2772; 29%) | Total PCR or rapid test for Flu A, Flu B, RSV (N = 41236) | p-value* | HAdV positive (N = 413; 39%) | HEV positive (N = 161; 15%) | MPV positive (N = 121; 11%) | Total PCR panel 2 (N = 1064) | p-value* | Total all viral assays (N = 42300) | |
| Age Mean (95%CI) | 4.6 (4.4, 4.9) | 2.8 (2.7, 2.9) | 2.9 (2.8, 3.1) | 3.296 (3.130, 3.462) | 3.2 (3.2, 3.3) | < 0.001 | 3.6 (3.5, 3.7) | 2.4 (2.3, 2.4) | 2.3 (2.2, 2.4) | 2.3 (2.2, 2.4) | 2.1 (1.9, 2.3) | 2.6 (2.6, 2.7) | 0.228 | Age Mean (95%CI) | 4.9 (4.8, 5.0) | 5.6 (5.4, 5.8) | 1.0 (0.96, 1.1) | 3.64 (3.61, 3.68) | < 0.001 | 3.0 (2.8, 3.2) | 2.8 (2.5, 3.1) | 2.8 (2.5, 3.1) | 3.1 (2.9, 3.2) | 0.084 | 3.6 (3.5, 3.6) | |
| Sex male | 378 (56.0%) | 722 (58.4%) | 954 (59.4%) | 456 (58.7%) | 2416 (58.4%) | 0.169 | 1027 (52.0%) | 1124 (59.9%) | 700 (56.9%) | 803 (58.7%) | 104 (58.8%) | 6134 (59.7%) | 0.453 | Sex male | 3736 (56.6%) | 979 (58.4%) | 465 (57.4%) | 23628 (57.3%) | 0.189 | 256 (62.1%) | 92 (57.5%) | 79 (65.3%) | 650 (61.2%) | 0.620 | 24127 (57.4%) | |
| ARI category: LRI | 612 (90.7%) | 909 (73.5%) | 1191 (74.2%) | 607 (78.1%) | 3035 (73.3%) | < 0.001 | 1545 (78.2%) | 1201 (64.0%) | 773 (62.8%) | 792 (57.9%) | 133 (75.1%) | 5890 (57.3%) | 0.607 | ARI category: LRI | 604 (9.1%) | 123 (7.3%) | 685 (84.6%) | 7463 (18.1%) | < 0.001 | 93 (22.5%) | 45 (28.0%) | 60 (49.6%) | 357 (33.6%) | < 0.001 | 8169 (19.4%) | |
| Co-detection | 298 (44.1%) | 606 (49.0%) | 660 (41.1%) | 777 (18.8%) | < 0.001 | 118 (6.3%) | 107 (8.7%) | 129 (9.4%) | 177 (1.7%) | Co-detection | 10 (0.2%) | 8 (0.5%) | 14 (1.7%) | 19 (0.0%) | < 0.001 | 98 (23.7%) | 70 (43.5%) | 53 (43.8%) | 130 (12.2%) | < 0.001 | 149 (0.4%) | |||||
a Allplex Respiratory Panel 4 (https://www.seegene.com/assays/allplex_respiratory_panel_4 ) using multiplex one-step real-time Polymerase Chain Reaction (PCR) of nasopharyngeal samples for the detection of seven bacteria causing respiratory tract infections, including Bordetella parapertussis (BPP), Bordetella pertussis (BP), Chlamydophila pneumoniae (CP), Haemophilus influenzae (HI), Legionella pneumophila (LP), Mycoplasma pneumoniae (MP), Streptococcus pneumoniae (SP). The table shows the patient characteristics for the total number of patients tested and the number of patients positive for each pathogen.
b Blood IgM serology test for Mycoplasma pneumoniae (LIAISON Mycoplasma pneumoniae IgM, Diasorin, Ireland). The table shows the patient characteristics for the number of patients with IgM seropositivity for Mycoplasma pneumoniae.
c Bacterial culture of nasopharyngeal samples (following the procedures guided in the “Clinical Microbiology Procedures Handbook” of the American Society for Microbiology (17)). The table shows patient characteristics for the total number of patients tested and the number of patients with positive cultures for each pathogen: Haemophilus influenzae (HI), Streptococcus pneumoniae (SP), Moraxella catarrhalis (MC).
d Viral assays including Allplex Respiratory PCR Panel 1 (https://www.seegene.com/assays/allplex_respiratory_panel_1 ) using multiplex one-step real-time RT-PCR for the detection of Influenza type A (Flu A), Influenza type B (Flu B) and Respiratory Syncytial Virus (RSV), or rapid test for the detection of Flu A and Flu B (SD Bioline Influenza A/B Ag of Standard Diagnostic, South Korea), and RSV (BD Veritor System RSV Devices of Becton, Dickinson and Company, USA) or Allplex Respiratory PCR Panel 2 (https://www.seegene.com/assays/allplex_respiratory_panel_2 ) using multiplex one-step real-time RT-PCR of nasopharyngeal samples for the detection of seven viruses causing respiratory tract infections, including Human Adenovirus (HAdV), Enterovirus (HEV), Metapneumovirus (MPV), Parainfluenza virus 1,2,3 4 (PIV1,2,3,4). The table shows the patient characteristics for the total number of patients tested and the number of patients positive for each pathogen.
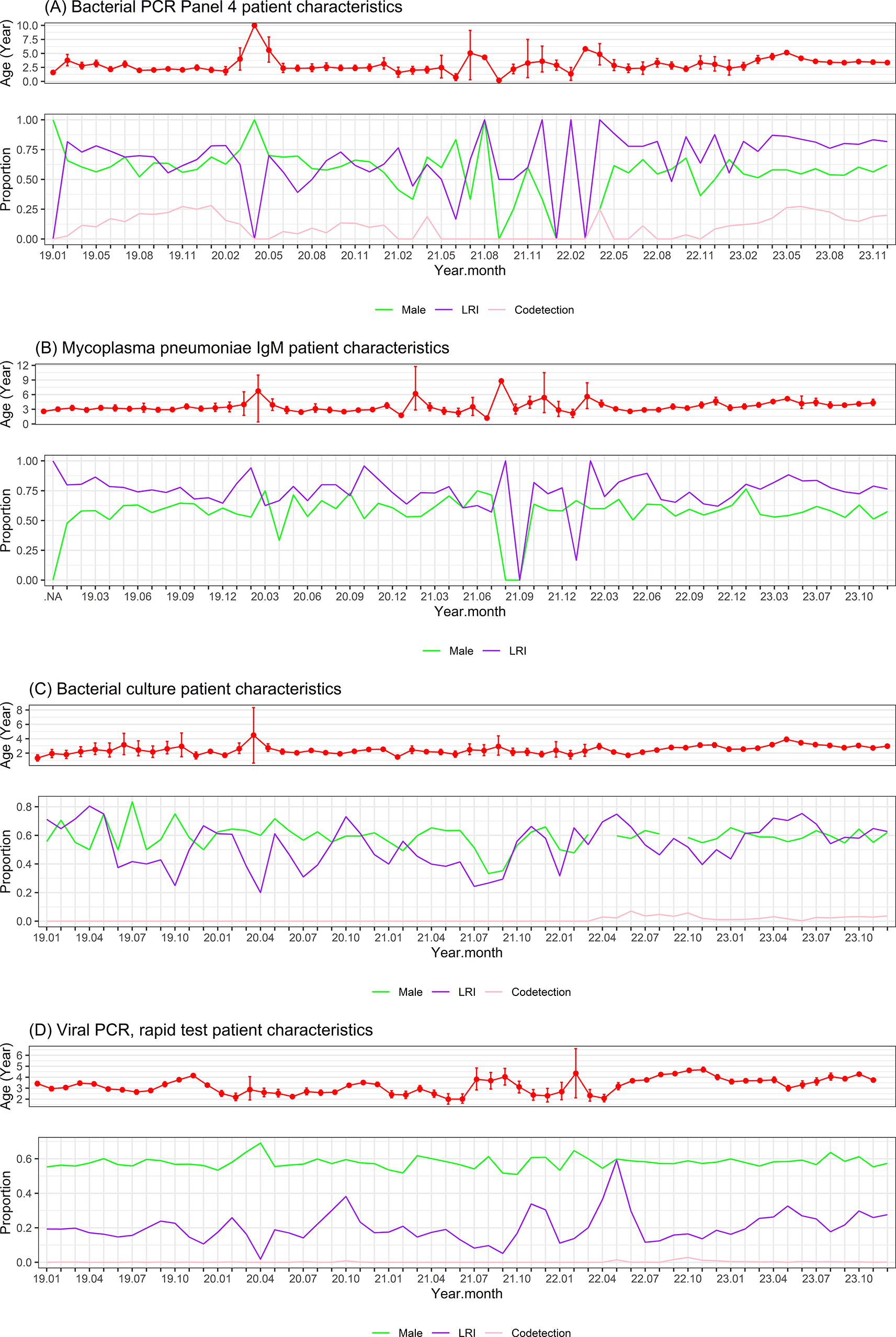
PCR, Polymerase Chain Reaction; LRI, Lower respiratory infection. Co-detection: ≥2 pathogens were detected. The x-axis shows the year and month of the year (e.g. 19.01 means January 2019, 23.10 means October 2023). The characteristics of all patients tested are displayed. Mean age with standard error, percentage of males, percentage of LRI, and percentage of co-detection were aggregated and plotted by month.
There were 5049 samples from 4361 patients tested for MP IgM, of which 1976 samples (39%) were serologically positive. The average age of those with seropositive MP IgM was 3.6 years old and 78% had LRI, and 52% were males ( Table 1, Figure 2B). There was no clear peak in MP IgM seropositivity before and during the COVID-19 pandemic from January 2019 to early 2022. In middle of 2022 when COVID-19 related restriction in Vietnam were relieved, even though there was an increase in the number of patients tested, the number of MP IgM seropositive cases remained low. By March 2023, a new peak of MP IgM seropositivity began to emerge in parallel with the peak of MP PCR positivity in 2023 ( Figure 1B).
Bacterial culture was performed on 10280 samples from 8073 patients, of which 4369 samples (43%) were positive for ≥1 bacterial pathogen. LRI accounted for 57%, males accounted for 60%, and the average age was 2.6 years old. The most common culture-positive pathogens were HI (N = 1878 (18%)), SP (N = 1230, 12%), and Moraxella catarrhalis (MC) (N = 1367, 13%). Co-detection of ≥2 pathogens was observed in 177 patients (1.7%). Co-detection was observed in approximately 6% of those positive for HI and 9% of those positive for SP or MC ( Table 1, Figure 2C). There was one peak of HI, SP, and MC infections from the end of 2020 to early 2021 when intermittent social distancing was applied, and there were only sporadic COVID-19 cases or clusters in Vietnam. After the relief of COVID-19 related restriction starting from the second quarter of 2022 in Vietnam, there was a large, long-lasting increase in positive bacterial cultures from the middle of 2022 until the end of the data period in December 2023 ( Figure 1C).
Regarding viral pathogens, Allplex PCR Panel 1 or rapid test for Flu A, Flu B, RSV, or Allplex PCR Panel 2 was done on 42300 samples from 28178 patients, of which 9777 (23%) samples were positive for ≥1 viral pathogens tested. A total of 6614/40112 (17%) samples were positive for Flu A, 1677/40112 (4%) samples were positive for Flu B, and 810/2772 (29%) samples were positive for RSV. In Allplex PCR Panel 2, 413/1064 (39%) samples were positive for HAdV, 161/1064 (15%) samples were positive for HEV, and 121/1064 (11%) samples were positive for MPV. LRI accounted for <10% of those positive for Flu A or Flu B, 85% of those positive for RSV, 23% of those positive for HAdV, 28% of those positive for HEV, and 50% of those positive for MPV. Co-detection was common in those positive for HAdV, HEV, or MPV (24–44%). Males accounted for approximately 60% of those positive for viral pathogens. The average age of those positive for HAdV, HEV, or MPV was approximately 3 years, the average age of those positive for RSV was 1 year, and the average age of those positive for Flu A or Flu B was approximately 5 years (p < 0.001) ( Table 1, Figure 2D). Flu A showed a regular cycle with peaks in the winter-spring months of 2019-2020, 2020-2021, 2022-2023 and the end of 2023 (except for a close-to-zero flat period during the lockdown due to the severe COVID-19 outbreak in the second half of 2021 to early 2022). Flu A peaks were often followed by smaller Flu B peaks a few months later. Soon after the relief of COVID-19 restriction starting from the second quarter of 2022, there was an exceptionally early peak of Flu A in the summer months of 2022, followed by a smaller peak of Flu B three months later. Another exception was the newly emerged peak of HAdV in the second half of 2022 ( Figure 1D).
The overall patient characteristics are summarized in Table 1 and plotted as a time series in Figure 2. Males accounted for ~60% of patients with ARI for all pathogens. Most bacteria-associated ARI patients had lower respiratory infections (LRI), whereas most virus-associated ARI patients (except RSV) had upper respiratory infections (URI). Most ARI patients were young (average age ~3 years old or younger), except those infected with MP, Flu A, and Flu B, who were slightly older (average age ~5 years old).
Our analysis of microbiology assay data of nasopharyngeal and blood samples of children with ARI from January 2019 to December 2023 provides an overall picture of the pattern of respiratory infections other than COVID-19 and the emerging pathogens before, during, and after the COVID-19 pandemic in Hanoi, Vietnam.
The reduction pattern of bacterial and other viral pathogens at the beginning of the pandemic from 2020 to early 2021 was somewhat similar to the pattern of bacterial and other viral pathogens in other countries such as Israel18 and China19 during this time. The close-to-zero flat period of bacterial and other viral pathogens during the lockdown period due to the severe COVID-19 outbreak in Vietnam in the second half of 2021 was explicable.
After the relief of COVID-19 related restriction starting from the second quarter of 2022 in Vietnam, the rebound of HI and SP was not only large in number of positive cases but also long-lasting. An increase in the number of HI-positive cases was observed from the middle of 2022 until the end of our data period in December 2023. Approximately three-fourths of these positive cases had LRI. This indicates that after the COVID-19 pandemic, HI and SP have largely re-emerged and become persistent epidemic pathogens associated with LRI in children. This pattern is similar to that observed in other countries, as summarized by Nygaard et al.20
After the relief of COVID-19 related restriction starting from the second quarter of 2022 in Vietnam, although the number of patients tested for MP IgM increased from early 2022, the number of patients with seropositive MP IgM did not increase until the middle of 2023. The increase in the number of patients with seropositive MP IgM was consistent with the increase in the number of patients with positive MP PCR results during the same period. This indicates that the increase was due to the real increase in the number of MP infection cases and not just due to the increase in the number of patients tested. Most of the patients positive for MP had LRI and were older than other patients positive for other bacterial pathogens. There was also a small newly emerged peak of CP-positive cases two months after the peak of MP-positive cases. These peaks were consistent with the outbreak of bacteria, especially atypical bacteria-associated pneumonia, in 2023, late after the COVID-19 pandemic, as reported in Vietnam,8 China,6 and some other countries.21
Overall, the pattern of viral pathogens during and after the COVID-19 pandemic in our data was quite similar to that in other countries, such as Italy9 and China.10–14 After the relief of COVID-19 related restriction starting from the second quarter of 2022 in Vietnam, the early rebound of Flu A and Flu B in the summer months of 2022 was consistent with the irregular rebound of viral ARI after the relief of COVID-19 restriction, as reported in many places.4,5 The exceptional newly emerged peak of HAdV in the second half of 2022 was consistent with the unprecedented outbreak of HAdV-associated ARI in children in Vietnam, as previously reported.7 A similar post-COVID-19 outbreak of HAdV-associated ARI in children was also reported a year later in Japan.22
Regarding the methodology, instead of plotting the positive rate, we plotted the number of samples tested and the number of samples positive for each pathogen by month. This would help contextualize the emergence or outbreak of pathogens better because it not only provides information on the number of samples tested and the number of positive samples (which are higher in case of outbreak) but also helps to infer the pathogen positive rate over time. Plotting the positive rates alone may be misleading. For example, one positive sample out of one tested sporadically would give a positive rate of 100%, but there would probably be no outbreak. Whereas 3000 positive samples out of 10000 samples tested in a short time would give a positive rate of 30%, and there would likely be an outbreak.
The strength of our study is the relatively large number of cases tested for various viral and bacterial pathogens over five years. This enabled us to examine the pattern and emergence of pathogens associated with ARI in children before, during, and after the COVID-19 pandemic. The main limitation of our study is that the data were from a single private hospital and might not accurately represent the overall population of children in Hanoi, Vietnam.
In brief, our data show that after the COVID-19 pandemic, HI and SP re-emerged as epidemic pathogens associated with LRI. Flu A and Flu B have re-established regular cycles of peaks in the winter-spring months after an early rebound, together with an unprecedented new HAdV emergence soon after the relief of COVID-19 restriction in 2022. Late after the COVID-19 pandemic, from the middle of 2023, the atypical pneumonia pathogen MP has emerged remarkably and has become epidemic, and there has also been a small, brief emergence of CP infection. Our data will be useful for infectious disease surveillance and public health intervention strategies in the post-COVID-19 era in Vietnam. Our data also provide additional and consistent useful information for health authorities to anticipate the impact of respiratory disease pandemics on the pattern of other pathogens associated with ARIs in children to prepare and manage if there is a similar pandemic in the future.
Repository: ‘STROBE checklist for Emerging pathogens associated with acute respiratory infections in children in Hanoi, Vietnam: An analysis of microbiology assay data from 2019 to 2023’ DOI: 10.5281/zenodo.1529387923
https://zenodo.org/records/15293879.
Data are available under the terms of the Creative Commons Zero ‘No rights reserved’ data waiver (CC0 1.0 Public domain dedication).
This study was approved (approval No 110/2024/CN/HDDD VMEC, date October 10, 2024) by the Ethical Committee of Vinmec Healthcare System and VinUniversity (Ethical Committee establishment decision No 24/2016/QD-VINMEC).
Informed consent was waived by the Ethical Committee for retrospective analysis and report of de-identified data.
The data generated and analyzed during the current study are not publicly deposited for security reason because they are protected property of the Vinmec Healthcare System. On reasonable data request for research or scientific validation purpose (not for commercial purpose and not for public data deposition), the data may be shared after data access application is approved by the Vinmec Ethical Committee and Vinmec Data Committee. Readers or reviewers may apply for access to the data by contacting the corresponding author via email: v.nhanht6@vinmec.com.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Jin R, Qin T, Li P, Yuan J, et al.: Increased circulation of adenovirus in China during 2023-2024: Association with an increased prevalence of species B and school-associated transmission. Journal of Infection. 2025; 90 (4). Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Metagenomics, Next-Generation Sequencing, Virology, Bioinformatics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 26 Dec 25 |
read | read |
|
Version 1 20 May 25 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)