Keywords
Diabetic Nephropathy, Mutations, Angiotensin Converting Enzyme Gene, Type 2 Diabetes Mellitus.
This article is included in the Genomics and Genetics gateway.
Diabetic nephropathy (DN) is a major complication of type 2 diabetes mellitus (T2DM) and a leading cause of kidney failure. Evidence on the influence of ACE I/D polymorphisms in DN risk is inconsistent across populations.
A systematic review and meta-analysis was conducted following the PRISMA 2020 guidelines. Studies published between January 1990 to February 2025 were retrieved from PubMed, EMBASE and Web of Science. Eligible observational studies reported the frequency of ACE genotypes with DN in T2DM. Independent reviewers screened studies using Rayyan software, extracted data, and assessed risk of bias using the ROBINS-E tool. Reporting on the quality of studies was determined using the STREGA guidelines. Pooled odds ratio (OR) with 95% confidence intervals (CI) were calculated using random-effects models in R version 4.4.2. Subgroup, meta-regression, and sensitivity analyses addressed heterogeneity; Egger’s test assessed publication bias. Registered in PROSPERO (CRD42024577680). Funding from Fogarty International Center of the National Institutes of Health (D43TWO11632).
Of the 46 studies included in this review, the combined sample size was 16,322 participants. The majority of studies (29 out of 46) were conducted in Asia. Only 5 studies reported DN–related comorbidities by ACE genotypes and one assessed mortality. Twenty-five of the included 46 studies contributed data to the meta-analysis. The ACE II genotype was protective against DN; II vs. ID [OR= 0.70 (CI: 0.63–0.77)] and II vs. DD [OR= 0.68 (CI: 0.55–0.84)]; Heterogeneity was (I2 = 71.7%, τ2 = 0.1776, p < 0.0001). Stronger associations were observed in studies using urinary Albumin-Creatinine-Ratio over Albumin-Excretion-Rate. Egger’s test showed no publication bias (p = 0.55).
The ACE II genotype is significantly protective against DN risk in T2DM. Standardization of urinary albumin measurement and further genotype-phenotype studies are needed to strengthen clinical utility of the ACE I/D polymorphisms.
Diabetic Nephropathy, Mutations, Angiotensin Converting Enzyme Gene, Type 2 Diabetes Mellitus.
Approximately 20–50% of people with diabetes worldwide suffer from diabetic nephropathy (DN), which is among the most prevalent and costly microvascular consequences of long-term type 2 diabetes mellitus (T2DM).1–3 Prolonged albuminuria, decreased glomerular filtration rate, and progressive kidney damage are hallmarks of DN, which eventually results in kidney failure.4 While microalbuminuria, commonly assessed via the urine albumin-to-creatinine ratio (UACR), remains a widely accepted early biomarker of diabetic nephropathy (DN),5 several other biomarkers have demonstrated potential for early detection. These include neutrophil gelatinase-associated lipocalin (NGAL), kidney injury molecule-1 (KIM-1), and cystatin C, which are often elevated before albumin appears in urine and may reflect early tubular injury or impaired glomerular filtration.6–8 Despite their clinical promise, these alternative biomarkers are not yet widely adopted in routine practice because of limitations such as high assay costs, limited availability of standardized kits, and the need for a specialized laboratory infrastructure.9,10 In contrast, microalbuminuria remains a practical and accessible option in most clinical settings, especially in low- and middle-income countries, owing to the affordability and availability of UACR testing.11–15
The pathophysiology of DN is complex, with hyperglycemia-induced reactive oxygen species, inflammation, and genetic predispositions playing pivotal roles.16,17 Angiotensin-converting enzyme (ACE), a key component of the renin-angiotensin-aldosterone system (RAAS), has been implicated in the development of DN because it is involved in the regulation of blood pressure and fluid balance. ACE gene insertion/deletion (I/D) polymorphism, which results in different genotypes insertion/insertion (II), insertion/deletion (ID) and deletion/deletion (DD), has been extensively studied as a potential genetic determinant of high blood pressure and DN risk.18–20 Evidence suggests that the ACE DD genotype is associated with increased ACE activity, leading to higher susceptibility to DN and its complications, whereas the II genotype may have a protective effect.21,22
Despite substantial research on diabetic nephropathy (DN), critical gaps remain in our understanding of the interplay between angiotensin-converting enzyme (ACE) insertion/deletion (I/D) genotypes and DN progression in type 2 diabetes mellitus (T2DM). While numerous studies have reported associations between ACE I/D polymorphisms and DN risk, the findings are inconsistent owing to variations in study design, population, and methods of microalbuminuria (UACR) measurement.23–26 The heterogeneity of these results could hinder the development of standardized clinical guidelines for the genetic risk assessment of DN.
Therefore, we conducted this systematic review and meta-analysis primarily to determine the association between Angiotensin Converting Enzyme (ACE) genotypes and Diabetic Nephropathy (DN) defined by Urine Albumin-to-Creatinine Ratio (UACR) in adults with Type 2 Diabetes Mellitus (T2DM). Secondary outcomes included DN-related mortality and comorbidities.
This systematic review and meta-analysis study was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.27 The protocol was registered with PROSPERO28 under registration number CRD42024577680 and is published as a Preprint in Open science Framework (OSF) preprints.29
Studies were eligible for inclusion if they enrolled male and female participants aged 18 years or older with a confirmed diagnosis of type 2 diabetes mellitus (T2DM), and investigated angiotensin-converting enzyme (ACE) insertion/deletion (I/D) genotypes (II, ID, DD) as the primary exposure.
Diabetic nephropathy (DN) was the primary outcome, commonly defined using microalbuminuria markers such as the albumin excretion rate (AER), urine albumin-to-creatinine ratio (UACR), or albumin-to-creatinine ratio (ACR). Diagnostic thresholds for microalbuminuria were standardized across studies at 30–300 mg/day for AER and 30–300 mg/g for UACR/ACR. One study that defined DN using traditional renal function tests, including estimated glomerular filtration rate (eGFR) and serum creatinine, was also included to capture broader clinical definitions of nephropathy. This deviation from the protocol was noted but deemed valuable for completeness.
The comparator across studies was typically T2DM patients with normal UACR levels, enabling evaluation of the association between ACE genotypes and DN risk. Secondary outcomes included nephropathy-related mortality and comorbidities such as cardiovascular disease, hypertension, neuropathy, and retinopathy. Eligible studies employed observational designs (cohort, case-control, or cross-sectional) and were published between January 1990 and February 2025.
Articles with missing data on key study variables or those that did not explore the ACE I/D-DN relationship were excluded. Non-English articles whose abstracts were available in English were included in the initial screening; however, those whose full texts were not available for retrieval were eventually excluded. Abstracts, unpublished studies, and grey literature were not included in this review, and no direct contact was made with study authors for clarification or additional data. Discrepancies were resolved through discussion, ensuring consistency and minimizing bias, although formal blinding and interrater reliability statistics were not applied.
We conducted a comprehensive search on across multiple electronic databases (PubMed, Web of Science, and EMBASE) to identify studies relevant to the research objectives. Additional studies were retrieved from Google Scholar and reference lists of the included articles to ensure broad coverage.
The search strategy was designed to capture studies assessing the relationship between angiotensin-converting enzyme (ACE) insertion/deletion (I/D) genotypes and diabetic nephropathy (DN). The search included studies published from 1st January 1, 1990, to 28th February 28, 2025, without language restrictions. Keywords and medical subject headings (MeSH) related to “ACE I/D genotypes,” “diabetic nephropathy,” and “microalbuminuria” were combined with Boolean Operators “OR” and “AND” “AND” and truncated where applicable to optimize retrieval. The search strings used are listed in Table 1.
Two independent experienced information retrieval specialists (AAK and ML), in consultations with the lead reviewer (RK), independently conducted the database searches and verified the inclusion of additional studies from alternative sources such as Google Scholar and reference lists. Discrepancies in the search results were resolved through consensus discussion. The systematic review process was organized using Rayyan,30 a web-based platform that facilitates the filtering and assessment of papers based on predetermined requirements for inclusion and exclusion.
Two reviewers, RK and JT, independently assessed the titles and abstracts of the studies according to the inclusion criteria using Rayyan software. Full-text articles were retrieved from studies that met the eligibility requirements. Any disagreements regarding inclusion were resolved through a consensus between the two reviewers. Full-text articles were retrieved from eligible studies, and the selection process is shown in Figure 1.
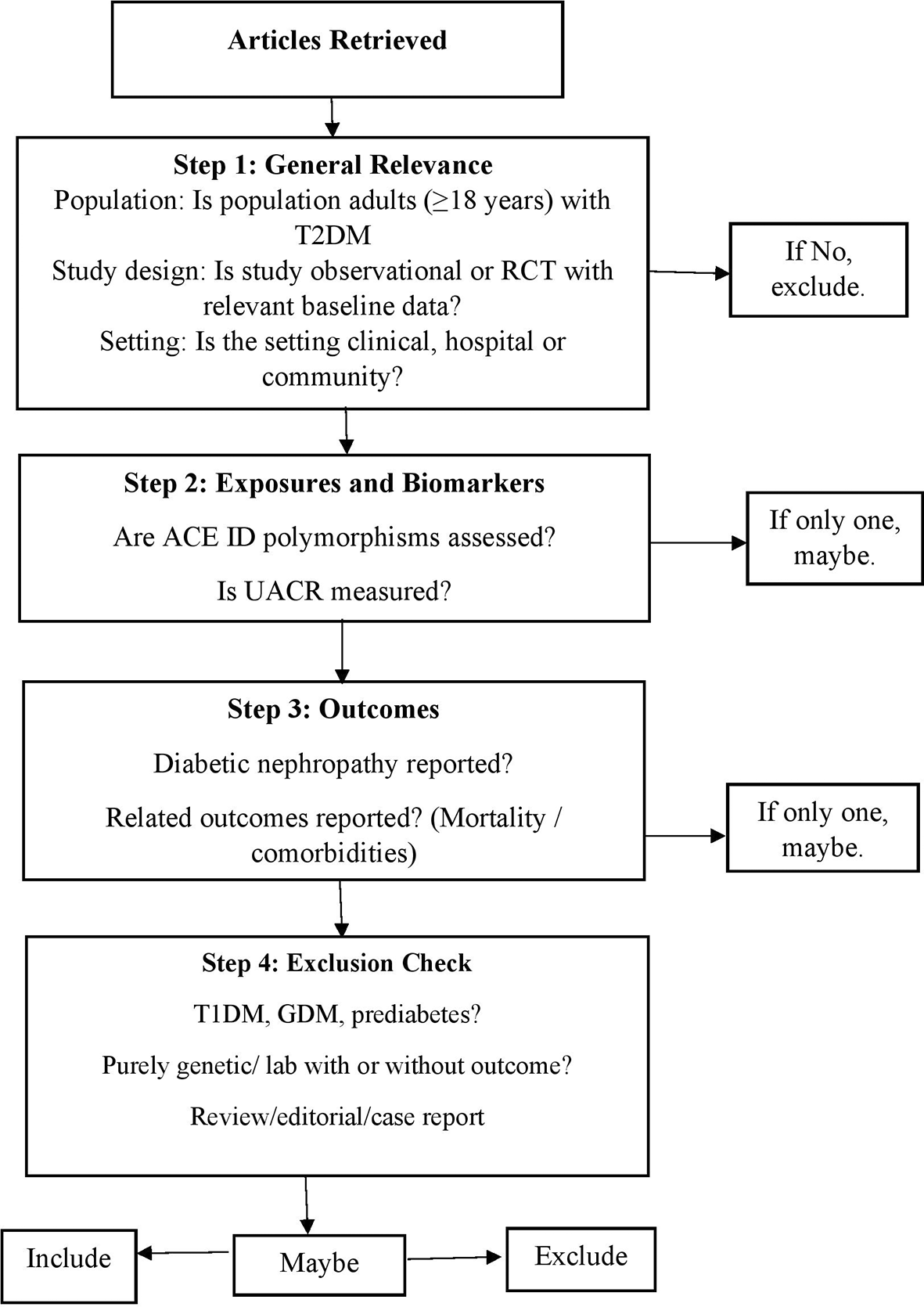
This flowchart illustrates the stepwise criteria used to screen and select articles for inclusion in the systematic review. The selection process involved four major steps: (1) assessing general relevance based on population, study design, and setting; (2) evaluating the presence of key exposures and biomarkers (i.e., ACE I/D polymorphisms and UACR); (3) confirming the reporting of diabetic nephropathy and related outcomes (e.g., mortality, comorbidities); and (4) applying exclusion criteria such as type 1 diabetes (T1DM), gestational diabetes (GDM), prediabetes, purely genetic/laboratory studies lacking clinical outcomes, and non-original articles (e.g., reviews or editorials). Articles were classified as “Include,” “Maybe,” or “Exclude” depending on how fully they met the criteria.
Abbreviations: T1DM – Type 1 diabetes mellitus; T2DM – Type 2 diabetes mellitus; GDM – Gestational diabetes mellitus; ACE – Angiotensin-converting enzyme; I/D – Insertion/deletion; RCT – Randomized Controlled Trial; UACR – Urine albumin-to-creatinine ratio.
Data extraction was performed independently by two authors, RK and JT, using a standardized data extraction form in Microsoft excel. The extracted data included study characteristics (author, year, population, and design, age, sex and clinical characteristics such as glycated hemoglobin levels, blood pressure, T2DM duration, body mass index), exposure details (ACE I/D genotypes), comparators (UACR measurement methods), outcomes (DN, mortality, and comorbidities), and statistical measures such as the frequency counts were also retrieved and computed in to Odds ratio with confidence intervals. Discrepancies were resolved through discussion with GNK and RA.
The data extraction captured a broad range of information including details on study design such as case control, cross-sectional or cohort, sample size and distribution between males and females, age, genotyping methods such as PCR, UACR or other DN diagnostic approaches, and outcome reporting. Genotype frequencies were entered in Excel, and odds ratio (OR) with 95% confidence intervals (CI) were calculated using R software version 4.4.2. Secondary outcomes such as comorbidities and mortality were recorded as binary variables (Yes/No) based on whether a particular study explicitly reported on them. No missing data were encountered during the meta-analysis, as only studies with complete and extractable data were included in the quantitative synthesis. Studies lacking sufficient data, as identified during visual inspection of the compiled Excel sheet, were excluded from meta-analysis and instead included in the narrative synthesis.
The risk of bias in individual studies was assessed using the ROBINS-E tool,31 which evaluates bias across key domains: confounding, selection bias, classification of interventions, and outcome assessment. Confounding was assessed as part of the ROBINS-E risk of bias evaluation, where considerations were made on whether studies appropriately controlled for potential confounders such as age, sex, duration of diabetes, and comorbidities. Where studies reported such adjustments or stratifications, these were documented and factored into the overall bias assessment. Studies were assigned an overall remark of either a low, moderate or some concerns, or high risk of bias. The risk of the bias assessments as depicted by the robvis tool32 are shown in Figure 2. The results of the bias assessment are available as Extended data. The ROBINS-E risk of bias assessment was conducted independently by CNB and SPR.
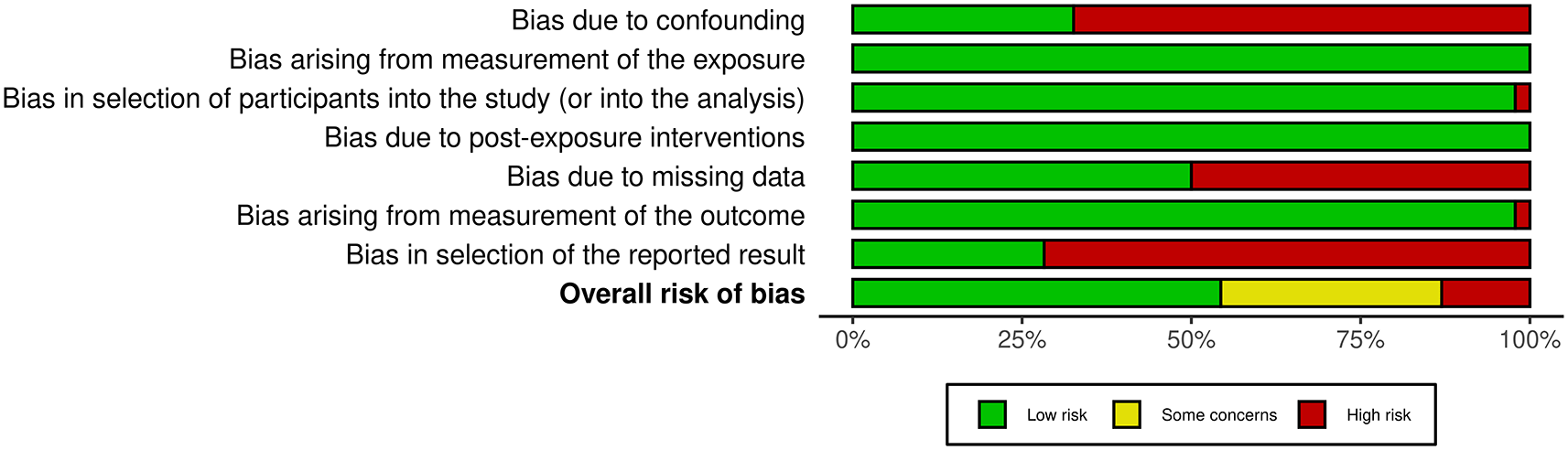
This bar plot illustrates the proportion of studies judged at each level of risk of bias across the seven ROBINS-E domains and overall, using the robvis tool. Most domains had a high proportion of low-risk ratings, particularly for confounding, exposure measurement, and participant selection. However, bias due to missing data and selective outcome reporting were the most frequent high-risk areas. In the “Overall risk of bias” domain, the bar plot shows approximately 60% of studies as low risk, which aligns with the actual count of 25 out of 46 studies (54%) that were included in the meta-analysis based on their low-risk rating.
We included 25 studies in the meta-analysis to assess the association between ACE I/D genotypes and diabetic nephropathy risk. For the primary outcome, diabetic nephropathy (DN) assessed using urine albumin to creatinine ratio or renal function tests, odds ratio (OR) with 95% confidence intervals (CI) were used as the effect measure to compare the risk of DN across ACE I/D genotypes (II vs. ID and II vs. DD). This binary outcome metric was selected due to the categorical nature of the genotype data and the case or non-case definition of DN. Odds ratio OR were calculated using genotype frequency data. Secondary outcomes such as comorbidities (hypertension, cardiovascular disease) and mortality were synthesized narratively, and their presence (Yes) or absence (No) was recorded per study.
Study selection and eligibility for synthesis
Studies were deemed eligible for synthesis if they included adult patients with T2DM, assessed ACE I/D polymorphisms and reported DN outcomes using laboratory-based criteria. The review included studies that assessed DN using albumin excretion rate (AER) or microalbuminuria, albumin-to-creatinine ratio (ACR), or renal function markers such as serum creatinine and estimated glomerular filtration rate (eGFR). These criteria allowed for flexible yet clinically relevant definitions of nephropathy. Study characteristics and outcome definitions were tabulated to determine synthesis eligibility. Studies without sufficient genotype data or outcome definitions were excluded.
Data preparation and conversion
Data preparation involved standardizing genotype frequency data across included studies. Frequencies for ACE I/D genotypes were extracted and used to calculate odds ratio (OR) with corresponding 95% confidence intervals (CI). For meta-analysis, all effect sizes were transformed into log odds ratios and back-transformed for interpretation. Secondary outcomes such as comorbidities and mortality were recorded as binary variables (reported vs. not reported) in Excel spreadsheets.
Reporting study outcomes
For the primary outcome, diabetic nephropathy (DN), the standardized metric used was the odds ratio (OR) with corresponding 95% confidence intervals (CI), comparing DN prevalence across ACE I/D genotypes (II, ID, DD). OR were chosen because they are appropriate for summarizing associations in observational studies. Genotype frequencies from individual studies were extracted into Microsoft Excel, and OR were calculated using R software (version 4.4.2). These OR were log-transformed for meta-analysis to stabilize variance and ensure normality assumptions. The pooled estimates were then back-transformed to the original OR scale.
For secondary outcomes, such as mortality, cardiovascular disease, hypertension, neuropathy, and retinopathy, data were extracted into Excel as binary indicators (Yes/No) depending on whether each study reported on the specific outcome.
Statistical models
The primary effect measure used was the odds ratio (OR) for the association between ACE I/D genotypes and DN. Random-effects models were used for all meta-analyses to account for between-study heterogeneity in population, study design, and UACR diagnostic methods. Statistical significance was determined at p < 0.05. Heterogeneity was assessed using the I2 and τ2 statistics. Meta-regression was performed to explore heterogeneity sources, particularly differences in urinary albumin measurement methods (ACR vs. AER). All statistical estimates were generated using R software (version 4.4.2).
Presentation of results
Forest plots were generated to visually present pooled OR and 95% CI. Individual study estimates and weights were shown. Funnel plots and Egger’s regression tests assessed publication bias. All statistical visualizations and analyses were produced in R software. Reporting bias was examined via funnel plots and Egger’s test; no significant asymmetry was detected.
Subgroup and sensitivity analyses
To explore sources of heterogeneity, subgroup analyses were conducted based on urinary albumin measurement methods (ACR vs. AER) of the included studies. Meta-regression was used to assess the potential influence of these study-level characteristics on effect size estimates.
Sensitivity analyses were conducted using a leave-one-out approach, in which each study was sequentially removed from the meta-analysis to evaluate the robustness and stability of the pooled effect estimates.
Selective outcome reporting and qualitative assessments were done through quality appraisal of the studies conducted using the STREGA reporting guidelines. KR and DT assessed adherence, with discrepancies resolved by ENO and BBA. Studies were rated as fair, good, very good, or excellent. Excellent studies provided thorough methodological and genotyping details; very good ones had minor gaps; good studies had moderate reporting deficiencies; and fair studies lacked core STREGA elements. The full results of STREGA appraisal are provided in the Extended data.
We did not apply the Grading of Recommendations, Assessment, Development and Evaluations (GRADE) framework,33 which is best suited for intervention studies. Instead, we appraised the certainty of evidence by triangulating risk of bias (via ROBINS-E), reporting quality of studies (via STREGA), consistency, precision of pooled estimates, and the direction of effects across studies.
The initial search across PubMed, EMBASE, Web of Science, Google Scholar, and reference lists yielded 968 records. After 189 duplicates were removed, 779 unique records were screened for eligibility. During the screening process, 721 records were excluded based on the title and abstract review. A total of 58 full-text articles were retrieved, all of which were successfully accessed. After full-text assessment, 12 studies were excluded due to incorrect population (n = 2), wrong outcome (n = 9), or wrong publication type (n = 1). Ultimately, 46 studies met the inclusion criteria and were included in this systematic review and meta-analysis. The complete screening and selection process is summarized in the PRISMA 2020 flowchart34 as shown in Figure 3.
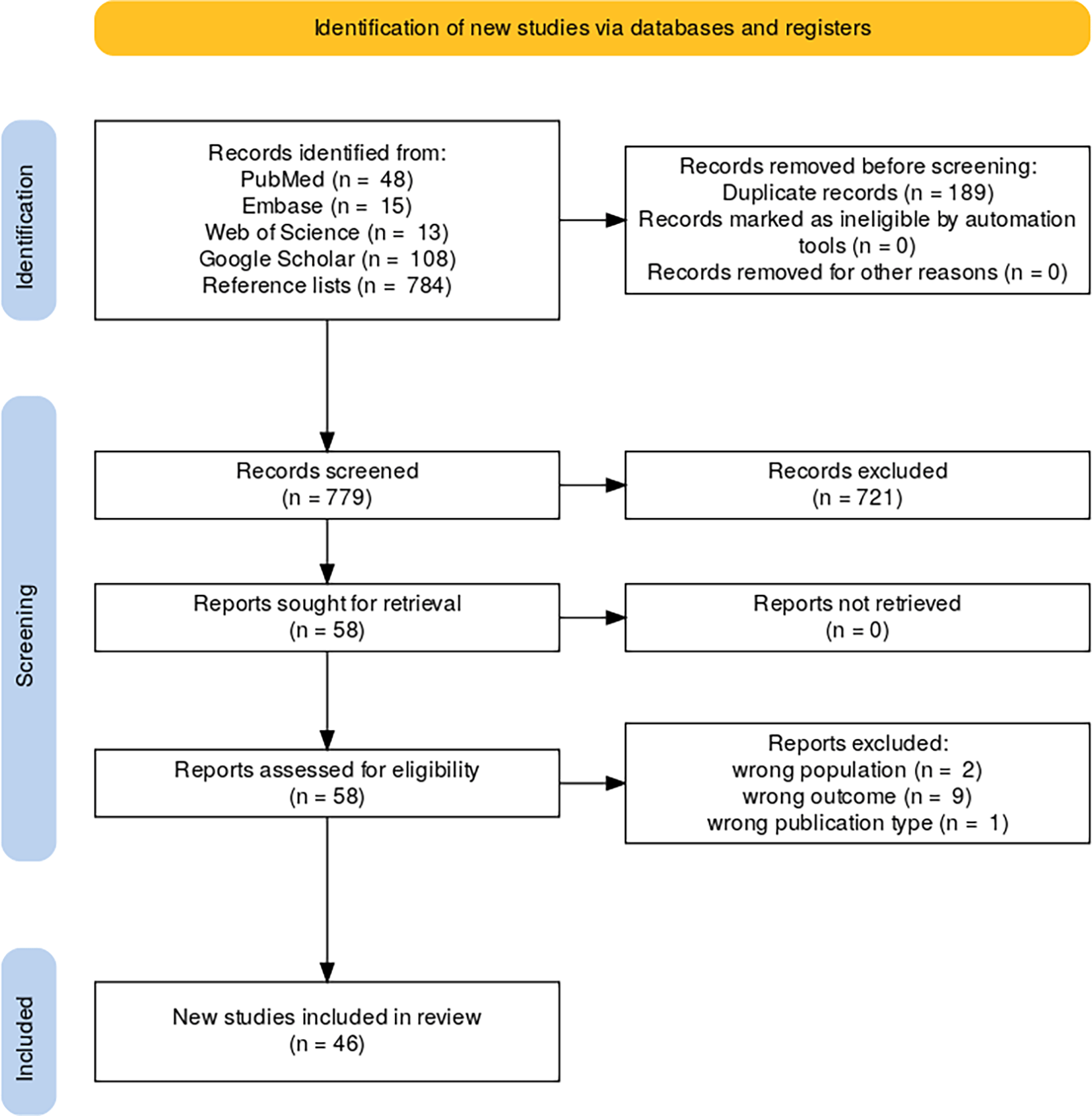
This flow diagram outlines the process of identifying, screening, and including studies in the review. A total of 968 records were identified through database searches and reference lists: PubMed (n = 48), Embase (n = 15), Web of Science (n = 13), Google Scholar (n = 108), and reference lists (n = 784). After removing 189 duplicates, 779 records were screened. Of these, 721 were excluded based on title and abstract screening, and 58 full-text reports were assessed for eligibility. Twelve reports were excluded due to; wrong population (n = 2), wrong outcome (n = 9), or wrong publication type (n = 1). Ultimately, 46 studies were included in the final review.
After full-text review, 12 studies that initially appeared to meet inclusion criteria were excluded for the following reasons: a) Wrong publication type (n = 1): Panagiotopoulos et al.,35 was a narrative review rather than a primary research study. b) Wrong outcome (n = 9): These studies, including Wong et al.,36 investigated ACE polymorphisms however, interest was with macroangiopathy and not type 2 diabetes mellitus associated nephropathy; Ahluwalia et al.,37 and Pai et al.,38 assessed ACE polymorphisms not as insertion/deletion SNPs. Fujisawa et al.,39 presented ACE I/D polymorphisms data in relation to retinopathy and myocardial infarction but not diabetic nephropathy (DN). Taha et al.,40 assessed ACE polymorphisms in type 2 diabetes mellitus patients but did not stratify them into nephropathy cases and controls. Araz et al.,41 ACE ID, DD, II were not distributed across UACR/albuminuric groups. Canani et al.,42 ACE DD and ID genotypes were not reported independently. Wong et al.,43 distribution of ACE I/D across different DN categories determined by UACR/Microalbuminuria/AER was not done. Draman et al.,44 microalbuminuria/UACR was not measured. c) Wrong Population (n = 2): Pontremoli et al.,45 focused on essential Hypertension and Al-Harbi et al.,46 in the methods section of the full text did not specify the population as type 2 diabetes mellitus (T2DM) but unrelated adult patients. These studies were excluded to ensure consistency with our predefined Population Exposure Comparator Outcome (PECO) criteria and focus on clinically relevant DN outcomes in adults with T2DM.
We followed the Centre for Reviews and Dissemination (CRD) approach47 and the Synthesis Without Meta-analysis (SWiM) guidelines48 due to their detailed and structured framework. Forty-six (46) observational studies investigating the association between ACE I/D polymorphisms and DN in patients with T2DM were examined. The total sample size across all studies was 16,322 participants, with individual study populations ranging from 50 to 3744 participants. The combined mean of mean age in the studies was 58.11 (±4.30). The combined median of mean numbers of males was 90.5 (IQR: 62.75-171.5) and that of females was 105.5 (IQR: 57.0-183.75). Most studies were conducted in Asia (29/46), Europe (8/46), Middle East (6/46) and Africa (5/46) reflecting a diverse geographical representation. Studies were grouped based on outcome assessment methods (ACR vs. AER), study design (case-control, cross-sectional, or cohort), and genotype comparisons (II vs. DD, II vs. ID), to account for methodological and clinical heterogeneity and to enable meaningful synthesis of effect estimates across similar contexts. Most studies assessed the primary outcome of DN using either ACR23,25,49–70 or AER,71–91 whereas only one study used traditional renal function tests92 (serum creatinine, eGFR, and urea levels). Genotyping for ACE I/D polymorphism was primarily conducted using polymerase chain reaction (PCR) methods, including conventional PCR, PCR-RFLP, high-resolution melting, and sequencing approaches. While all studies reported ACE I/D genotype distributions and DN outcomes, only five studies (10.7% of 46) explored associations with comorbidities, and one (2.2% of 46) reported on mortality. The majority identified either the D allele or DD genotype as being associated with increased DN risk, although several (40/46) reported null or population-specific findings.
The geographical distributions of the studies included in this review are shown in Figure 4.
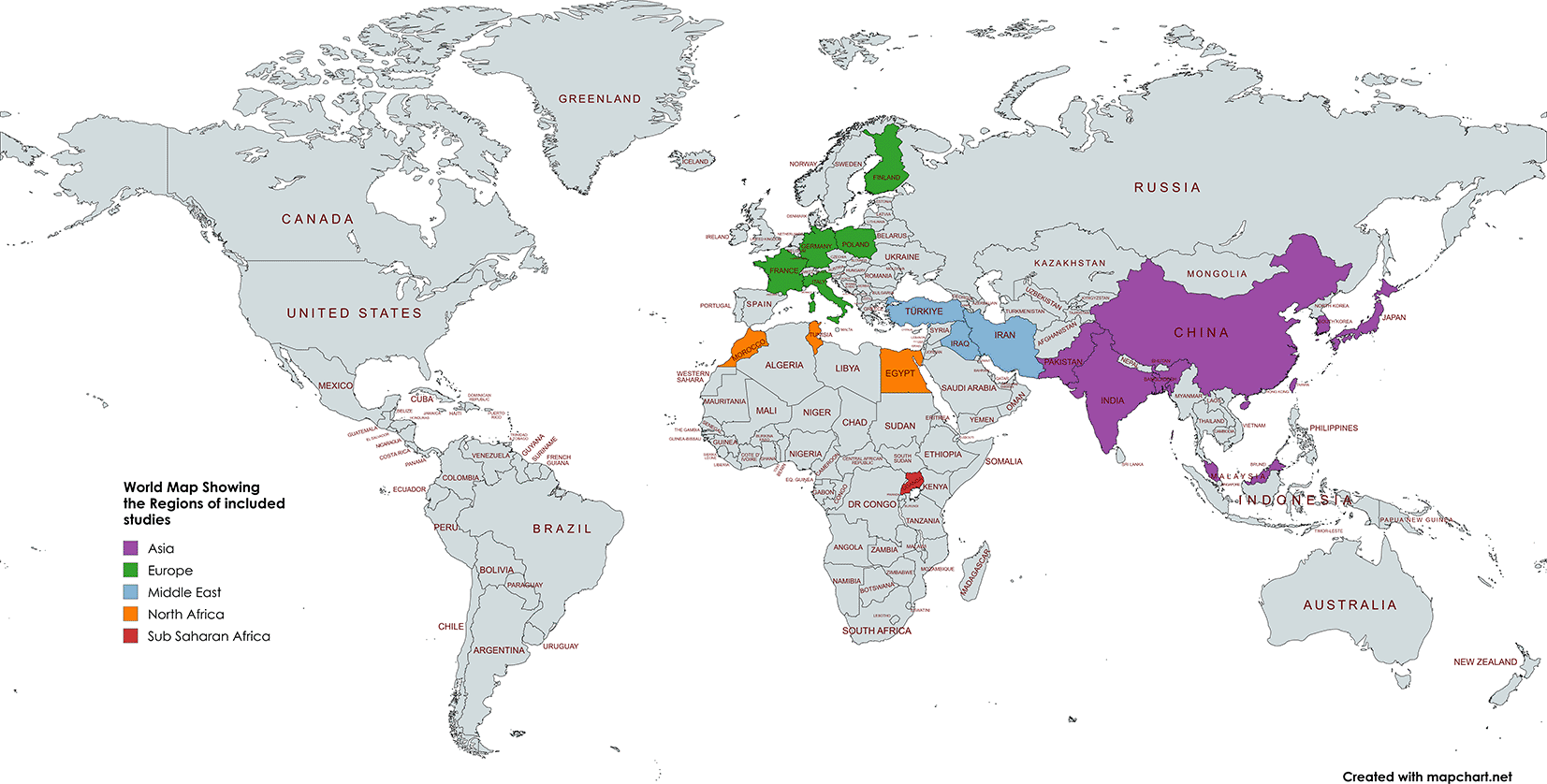
This world map illustrates the countries where the 46 studies included in the systematic review were conducted. The studies are distributed across several regions, including Asia (China, Japan, South Korea, India, Pakistan, Taiwan, Bangladesh, Malaysia), the Middle East (Iran, Iraq), Europe (Finland, France, Germany, Italy, Poland, Turkey), and Africa (Egypt, Morocco, Tunisia, Uganda). Countries are color-coded to reflect regional groupings. The map was generated using MapChart.net.
Due to the limited number of studies addressing ACE I/D polymorphism in relation to diabetic nephropathy and associated comorbidities, the included studies were synthesized narratively rather than statistically. The summary of findings examining ACE I/D polymorphism in relation to diabetic nephropathy and comorbidities is presented in Table 2.
From the 46 studies, only those with a low overall score on the ROBINS E assessment were included in the quantitative synthesis. We conducted a meta-analysis, meta-regression, subgroup analysis, sensitivity analysis, and publication bias assessment on 25 studies to evaluate the association between ACE I/D polymorphisms and diabetic nephropathy (DN) in the study population.
This was conducted in accordance with the Meta-analysis of Observational Studies in Epidemiology (MOOSE) guidelines,93 and the corresponding checklist was completed to ensure comprehensive and transparent reporting. The included studies were deemed appropriate for assessing the hypothesis as they reported on ACE I/D genotypes and diabetic nephropathy outcomes in adult T2DM populations. Data were selected and coded based on sound clinical principles, prioritizing studies that clearly defined diabetic nephropathy using established albuminuria or renal function markers and reported ACE I/D genotype distributions relevant to the study objectives. Data classification and coding were performed independently by two reviewers using a standardized extraction template. Most included studies used standardized measures of albuminuria to define nephropathy, ensuring relevance to the primary outcome and alignment with the review objective. Data classification and coding were performed independently by two reviewers (RK and JT) using a standardized extraction template. Discrepancies were resolved through discussion with another reviewer; EAO, ensuring consistency and minimizing bias, although formal blinding and interrater reliability statistics were not applied.
Random-effects models using the DerSimonian–Laird method94 were used to estimate the pooled odds ratios (ORs) and 95% confidence intervals (CIs) across genotype comparisons to account for expected heterogeneity in study populations and methodologies across the included observational studies. Genotype II was associated with a lower risk of developing DN. The random-effects models showed a significant protective effect of the homozygous insertion (II) genotype, with pooled odds ratio (OR) of 0.70 (95% CI: 0.63–0.77) versus heterozygous insertion/deletion (ID) and 0.68 (95% CI: 0.55–0.84) versus homozygous deletion (DD). Heterogeneity across studies was moderate to substantial (I2 = 71.7%, τ2 = 0.1776, p < 0.0001), suggesting variability in the effect estimates, possibly due to methodological or population differences. A forest plot summarizing the pooled estimates and heterogeneity is shown in Figure 5.
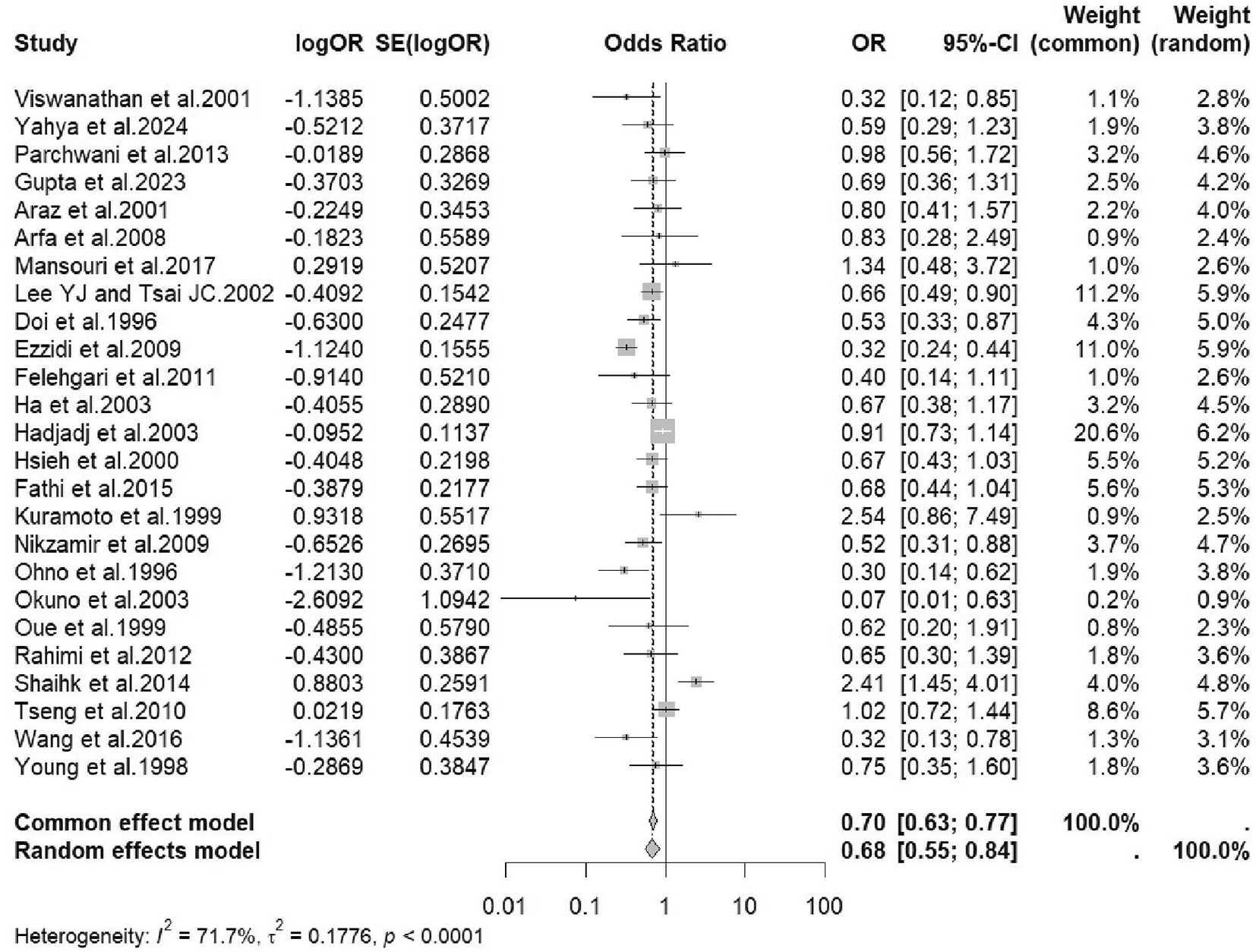
This forest plot presents the individual and pooled odds ratio (OR) with corresponding 95% confidence intervals (CIs) for studies included in the meta-analysis assessing the association between ACE I/D polymorphism and the risk of diabetic nephropathy. Each horizontal line represents the confidence interval for a single study, with the square marker indicating the point estimate of the OR. The size of each square reflects the weight assigned to the study in the random-effects model. The diamond at the bottom of the plot represents the overall pooled OR, with a value of 0.68 (95% CI: 0.55–0.84). The heterogeneity among studies was moderate (I2 = 71.7%, p < 0.0001).
Meta-regression was performed to explore the sources of heterogeneity in the association between ACE I/D genotypes and diabetic nephropathy (DN) risk. In the crude model (k = 25; DerSimonian–Laird estimator), the residual heterogeneity was τ2 = 0.1100 (SE = 0.0917; τ = 0.3317), with I2 = 52.3% (p = 0.0141) and R2 = 38.06%. The test for residual heterogeneity was significant (QE(12) = 25.16, p = 0.0141), while the test for moderators was not (QM(12) = 16.56, p = 0.17), indicating limited explanatory power overall. However, the form of UACR measurement was a significant individual predictor (β = 0.57, SE = 0.19, p = 0.0033; 95% CI: 0.19-0.95), suggesting that variations in measurement methods contributed meaningfully to heterogeneity in DN risk estimates. In the adjusted model, which included additional moderators, the residual heterogeneity increased (τ2 = 0.1253, SE = 0.0692; τ = 0.3540), with I2 = 63.0% (p < 0.0001) and R2 = 29.45%. The overall test for residual heterogeneity remained significant (QE(23) = 62.21, p < 0.0001). The test of moderators was significant (QM(1) = 5.79, p = 0.016), confirming that the form of UACR measurement remained a key source of heterogeneity (β = 0.40, SE = 0.17, p = 0.016; 95% CI: 0.07-0.72). The results are summarized in Table 3.
Subgroup analyses were based on the methods used to assess the primary outcome which is DN and these included; ACR, AER and RFTs. They revealed differences in the strength of associations between ACE I/D genotypes and diabetic nephropathy (DN) risk. Studies using albumin-to-creatinine ratio (ACR) showed the strongest protective effect of the II genotype, with pooled odds ratio (OR) of 0.59 (95% CI: 0.47–0.75) under the random-effects model and 0.59 (95% CI: 0.51–0.67) under the common-effect model. For studies using the albumin excretion rate (AER), the association was weaker (random-effects OR = 0.71, 95% CI: 0.55–0.92; common-effect OR = 0.77, 95% CI: 0.66–0.90). The weakest association was observed in the study using renal function tests (RFTs), with a random-effects OR of 0.68 (95% CI: 0.55– 0.84) and a common-effect OR of 0.70 (95% CI: 0.63–0.77). Heterogeneity varied across subgroups: I2 = 60.3% (p = 0.0019) for ACR and I2 = 58.3% (p = 0.0102) for AER; no heterogeneity was detected in the RFT-based subgroup. These results suggested that the method used to assess DN may influence the estimated association between ACE I/D polymorphisms with DN risk. The full summary of subgroup results is presented in Table 4.
Leave-one-out sensitivity analysis was conducted to assess the robustness of the findings and the influence of individual studies on the pooled effect estimates. One study was excluded at a time, and the overall odds ratio (OR) and 95% confidence interval (CI) were calculated. The pooled log OR ranged from -0.34 to -0.45, with minimal variation across iterations. The confidence intervals remained narrow and did not cross the null, suggesting consistent associations. The exclusion of Shaihk et al.92 produced the most marked change (log OR = -0.4474; 95% CI: -0.6255 to -0.2692), followed by Kuramoto et al.81 and Tseng et al.,66 though none altered the overall interpretation. These results indicated that no single study had a disproportionate impact on the association between ACE I/D genotypes and diabetic nephropathy in the meta-analysis. The full results are shown in Table 5 and visualized in Figure 6.
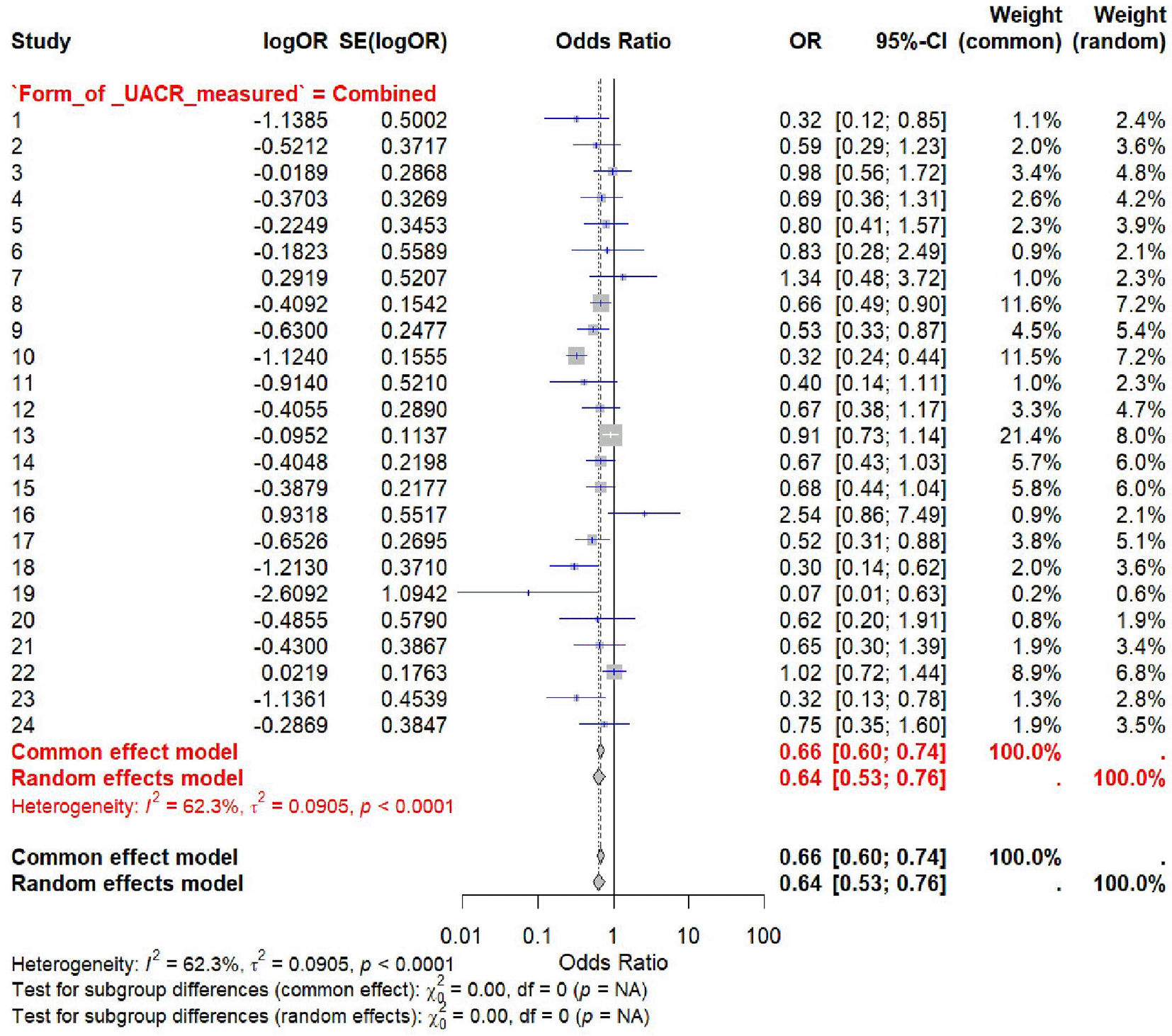
This forest plot illustrates the results of a leave-one-out sensitivity analysis. Each line represents the pooled odds ratio (OR) and 95% confidence interval (CI) calculated by excluding one study at a time from the overall analysis. The consistency of effect estimates across iterations indicates that no single study unduly influenced the pooled results. The overall pooled OR remains significant and stable (OR = 0.64; 95% CI: 0.53–0.76) under the random-effects model. Heterogeneity was moderate (I2 = 62.3%, p < 0.0001), and the form of UACR measurement was included as a subgroup moderator.
Abbreviations: ACE – Angiotensin-Converting Enzyme; I/D – Insertion/Deletion; OR – Odds Ratio; CI – Confidence Interval; SE – Standard Error; UACR – Urine Albumin-to-Creatinine Ratio.
The funnel plot shown in Figure 7 and Egger’s regression test were used to assess the publication bias. The plot shows a relatively symmetrical distribution around the pooled effect size, with no substantial asymmetry. Egger’s test (t = -0.61, df = 23, p = 0.55; bias = -0.50, SE = 0.82) indicated no significant small study effects. The residual heterogeneity variance was τ2 = 3.62, using standard error as the predictor and inverse-variance weights. Given the lack of asymmetry, a trim-and-fill analysis was not performed. These results suggest that the pooled estimates are unlikely to be affected by publication bias.
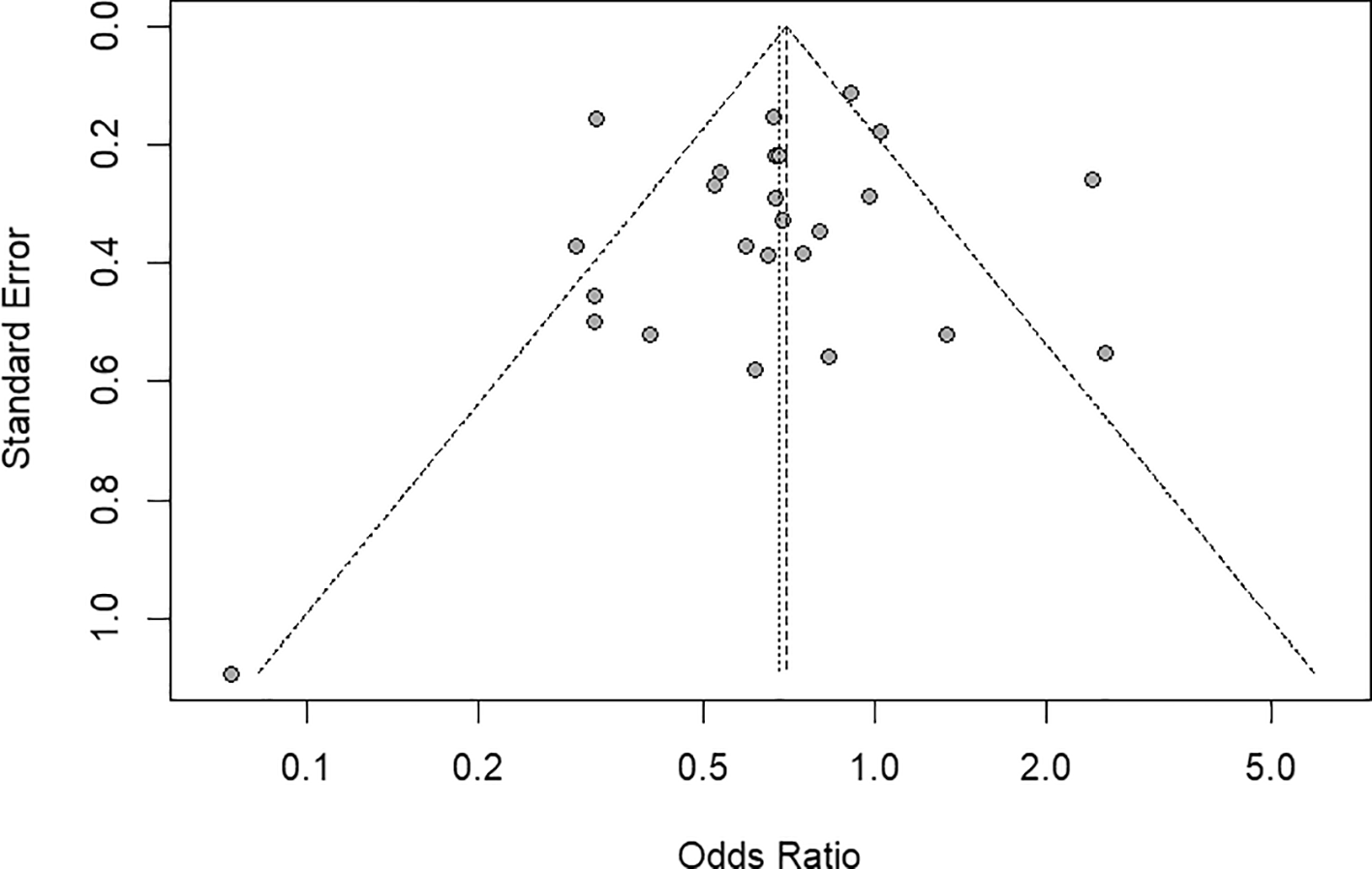
This funnel plot visualizes the distribution of studies included in the meta-analysis. The plot displays the standard error (y-axis) against the odds ratio (x-axis) for each study. Symmetry of the scatter around the pooled effect estimate suggests the absence of significant publication bias. The majority of studies fall within the pseudo 95% confidence limits (dashed lines).
Abbreviations: ACE – Angiotensin-Converting Enzyme; I/D – Insertion/Deletion; OR – Odds Ratio; SE – Standard Error.
Assessment of reporting bias was informed by the “Selection of Reported Result” domain in ROBINS-E and several reporting items in STREGA. In ROBINS-E, most studies were rated as having a moderate risk for selective reporting, indicating that not all measured outcomes may have been reported. In the STREGA assessment, most studies did not report sample size justification or apply multiple testing correction, and Hardy-Weinberg Equilibrium (HWE) was tested in only a few. These findings suggest a moderate risk of bias due to missing or selectively reported results across the included studies.
The certainty of the evidence in this review was appraised qualitatively, as the GRADE approach was not applied due to the observational nature of the included genetic association studies. Instead, confidence in the findings was supported by several methodological safeguards: risk of bias was assessed using the ROBINS-E tool, guiding the inclusion of only low-risk studies in the meta-analysis; reporting quality was evaluated using the STREGA checklist to ensure transparency and reproducibility; and consistency, direction, and precision of effect estimates were examined across studies. Out of 46 studies, 25 were rated as having low risk of bias, 15 as moderate or some concerns, and 6 as high according to ROBINS-E. STREGA compliance ratings showed that 19 studies were rated as very good, 13 as fair, 9 as good, and 5 as excellent. Based on this distribution, the overall confidence in the evidence is moderate, reflecting generally sound methodology and reporting, though limitations in transparency and bias remain in a subset of studies.
Heterogeneity was assessed using I2 and τ2 statistics and explored further through subgroup and meta-regression analyses. Publication bias was evaluated through funnel plots and Egger’s regression test. Together, these assessments provide a moderate to high level of confidence in the observed association between ACE I/D genotypes and diabetic nephropathy, particularly the protective effect of the II genotype. However, the certainty of evidence for secondary outcomes, such as mortality and comorbidities, remains low due to limited data.
We evaluated the association between ACE insertion/deletion (I/D) polymorphisms and DN in T2DM patients, with a focus on differences in UACR assessments, study populations, and clinical outcomes such as hypertension, cardiovascular disease, stroke and mortality from DN. Narrative synthesis demonstrated a generally consistent association between ACE I/D variants and DN risk, although the effect sizes varied across studies. The extent to which ACE I/D genotypes influence DN outcomes likely varies depending on several intersecting factors. Population characteristics, such as geographical location, age, sex, and prevalence of hypertension or other metabolic comorbidities, which modify the genetic effect of the ACE I/D polymorphism. For example, the distribution of I/D alleles and their association with DN may differ among Asian, African, and European populations due to underlying genetic diversity and environmental exposure.
The meta-analysis revealed that individuals with the ACE II genotype had significantly lower odds of developing diabetic nephropathy compared to those with ID or DD genotypes, highlighting a protective genetic association as the primary outcome of this systematic review. Previous studies that encompassed ethnicity on this topic found that there was a reduced risk of diabetic nephropathy associated with genotype II among Europeans (Caucasians) with type 2 diabetes, although this observation was more applicable to Asians (Chinese, Japanese, Koreans).22,95 Moreover, the meta-analysis confirmed a statistically significant protective association of the II genotype compared to both ID and DD genotypes, consistent with prior research implicating the renin–angiotensin–aldosterone system (RAAS) in DN pathogenesis.22,56,95 These findings reinforce the hypothesis that genetic variation at the ACE locus contributes to an individual’s susceptibility to DN. However, the modest magnitude of the effect suggests that this polymorphism is likely one of multiple genetic contributors within a polygenic framework, as noted from the heterogeneity in the method used to assess diabetic nephropathy in primary studies.
We observed that the identification of DN cases varied considerably across studies. Most studies assessed DN using either ACR or AER, while only one study relied on traditional renal function tests, such as serum creatinine, estimated glomerular filtration rate (eGFR), or urea levels. This inconsistency in diagnostic criteria significantly affects the comparability and generalizability of reported genotype-phenotype associations. Moreover, in our meta-regression analysis, the UACR measurement method emerged as a critical moderator of heterogeneity, underscoring the methodological influence on the pooled effect sizes. Although these biochemical markers are practical for large-scale research and clinical use, they are surrogates rather than definitive diagnostic tools. Notably, kidney histology remains the gold standard for diagnosing DN, offering detailed insight into glomerular and tubular changes.96 However, renal biopsy is rarely performed due to its invasive nature, associated risks, and ethical limitations in asymptomatic populations.97
Subgroup analyses revealed stronger genotype-phenotype associations in studies using urinary albumin-to-creatinine ratio (UACR) compared to albumin excretion rate (AER) or serum-based renal function tests. These findings align with prior recommendations advocating standardized albuminuria assessment in DN research to improve reproducibility and comparability across studies.98,99 Methodological inconsistency in DN diagnosis remains a major source of heterogeneity in the genetic literature.100 These methodological differences may obscure or exaggerate the true effects of ACE I/D polymorphism.101,102
Sensitivity analyses confirmed the robustness of the pooled estimates, with no individual study unduly influencing the overall results. While studies by Shaikh et al.,92 Kuramoto et al.,81 and Tseng et al.66 showed relatively greater influence when excluded, the overall direction and significance of the associations remained unchanged. This stability across leave-one-out analyses supports the internal validity of the meta-analysis. Assessment of publication bias using funnel plot symmetry and Egger’s regression test indicated no evidence of small-study effects.103 The non-significant test and symmetrical funnel distribution suggest that selective reporting did not materially influence the meta-analysis findings.
Despite the breadth of studies evaluating the genetic associations with DN, a major gap exists in the reporting of clinical outcomes. From the narrative synthesis of the included studies in this review, only one study addressed DN-related mortality,83 and five reported comorbidities such as metabolic disease,104 neuropathy56 and retinopathy.52,57,70 Most studies have been conducted in broader T2DM populations without disaggregating the outcomes for DN-specific cohorts. Consequently, the translational relevance of ACE I/D polymorphisms to clinical endpoints in DN remains poorly defined. Future studies should prioritize DN-specific outcomes and adopt standardized reporting frameworks to improve clinical interpretability.
One limitation in this study is the presence of moderate between-study heterogeneity. This heterogeneity persisted even after subgroup and meta-regression analyses were conducted. A likely explanation for this is residual confounding. This may be due to variations in ethnicity, environmental exposure, and gene–environment interactions. Unfortunately, these variables were not captured in the data-extraction process. For instance, although the review predominantly included studies defining diabetic nephropathy based on urinary albuminuria markers (ACR or AER), one study using serum creatinine and eGFR was retained to incorporate relevant genetic data. This slight deviation was carefully considered and does not substantially compromise the consistency or validity of the overall findings, as the included outcome remains clinically relevant to nephropathy assessment. In addition, the primary diagnostic marker for DN in our systematic review was urinary albumin-to-creatinine ratio (UACR), as reported in the original studies. However, we acknowledge that there are differences between the quantitative and semi-quantitative methods for measuring urinary albumin levels. To address this, we used the ROBINS-E tool to assess the risk of bias related to outcome measurements. Studies that lacked clarity or likely used semi-quantitative methods were rated as moderate-to-high risk, and only those with low overall risk (25 studies) were included in the meta-analysis. This helped ensure a more accurate DN classification in our pooled estimates. Moreover, in our subgroup/meta-regression analyses, the form of urinary albumin measurement; ACR vs. AER was statistically significant, reinforcing the impact of methodology on DN diagnosis. This review did not include unpublished studies or abstracts, and no author contact was made for missing data, which may have limited access to additional insights; however, efforts were made to ensure comprehensive coverage through rigorous database and reference list searches.
Therefore, we suggest that future studies prioritize standardized methods for assessing microalbuminuria, particularly the use of ACR over AER, to reduce heterogeneity in outcome reporting. There is also a clear need for well-designed, large-scale genetic association studies that specifically examine ACE I/D polymorphisms in relation to DN-related mortality and comorbidities, such as hypertension and cardiovascular disease, which are underrepresented in the current literature. Additionally, future research should explore gene-environment and gene-gene interactions, as well as the utility of ACE genotyping in clinical risk prediction models for DN progression within diverse populations, especially in regions such as sub-Saharan Africa, where such data are scarce.
This systematic review and meta-analysis provides evidence for a protective association between ACE insertion/deletion (I/D) polymorphism and diabetic nephropathy (DN), with genotype II consistently linked to reduced DN risk. Subgroup and meta-regression analyses identified the method of urinary albumin measurement; in particular, the albumin-to-creatinine ratio (ACR), which, according to our study, was a significant moderator of effect size variability, highlighting the need for methodological standardization in DN research and clinical practice. These findings were robust to sensitivity testing and showed no evidence of publication bias. However, the limited reporting of DN-specific clinical outcomes, including mortality and comorbidities, highlights persistent gaps in the literature. Moderate heterogeneity across studies suggests additional contributing factors; therefore, future research should prioritize harmonized diagnostic criteria, expand the evaluation of clinical outcomes, and incorporate polygenic and environmental risk models. Such efforts are essential for advancing precision medicine approaches for the prevention and management of DN in individuals with type 2 diabetes.
This study is part of a doctoral research project approved by the Mbarara University of Science and Technology Research Ethics Committee (MUST-REC), reference number MUST-2024-1725. The review was conducted using publicly available data and therefore did not require additional ethical clearance.
RK - Ritah Kiconco, KR- Robert Kalyesubula, RA- Raymond Atwine, JT- Jazira Tumusiime, CNB- Charles Nkubi Bagenda, SPR- Simon Peter Rugera, BBA- Bosco Bekiita Agaba, ENO- Erick Nyakundi Ondari, DT- Deusdedit Tusubira, GNK- Gertrude N. Kiwanuka, ML - Martha Lyaka, MO - Moses Ocan, AAK- Alison Annet Kinyengere, EO - Ekwaro Obuku.
The materials used in this review are available in Zenodo. The project title is “ACE I/D gene mutations in diabetic nephropathy” https://doi.org/10.5281/zenodo.15765008105
The project contains the following extended data;
• Completed Table 2
• 2020 PRISMA checklist
• MOOSE checklist
• SWiM checklist
• ROBINS-E assessment
• STREGA assessment
• Compiled meta-analysis dataset
• Compiled systematic review dataset
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The authors acknowledge the support of Mbarara University of Science and Technology (MUST) and the Africa Centre for Systematic Reviews and Knowledge Translation for providing technical and academic guidance throughout the development of this review. We thank the Fogarty International Center of the NIH for supporting the lead author through the D43 multimorbidity research training grant. The funder had no role in this review’s design or publication. Special thanks to all collaborators and institutional mentors who provided feedback and oversight throughout the reporting process.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)