Keywords
COVID-19; idiopathic hepatitis; population health; liver chemistries; pandemic
This article is included in the Coronavirus (COVID-19) collection.
SARS-CoV-2 infection is novel and widespread globally, causing a range of illness severity. The impacts on the liver from SARS-CoV-2 are variable and appear unpredictable.
In the weeks following SARS-CoV-2 infection, liver-related blood tests are abnormal in patterns different than in matched controls without SARS-CoV-2 infection.
This was a retrospective study of health administrative data of individuals in Ontario, Canada between January 2020 and September 2022. Cases were individuals with their first positive COVID-19 test with at least one liver-related blood test (AST, ALT, albumin, INR, bilirubin) available in the following 28 days. Two control individuals (with a negative COVID-19 test) were age/sex matched to each case. Any patients with a diagnostic code indicting pre-existing liver disease, or a positive COVID-19 test in the preceding six months were excluded. Machine learning analysis approaches included logistic regression, random forest, extreme gradient boost (XGBoost), and neutral network models.
100,574 cases and 200,709 matched controls were included. 49% (49,299) had at least one abnormal liver test in 28 days following positive COVID-19 test. Of the controls, 41% (82,495) had at least one abnormal liver test in the 28 days following negative COVID-19 test. The optimal machine learning model used XGBoost for adult cases, with an area under the curve of 0.74.
Further studies could use this approach to create monitoring systems to use abnormalities like liver enzyme changes, at a population level, as a bellwether for not only COVID-19 outbreaks, but also other hepatotropic illness epidemics, and pandemics.
COVID-19; idiopathic hepatitis; population health; liver chemistries; pandemic
• The impacts on the liver from SARS-CoV-2 are variable and appear unpredictable.
• This study leverages over 300,000 individuals who had liver and COVID-19 tests.
• A subset of individuals with COVID-19 had elevated ALT, AST, and lower albumin.
• The transaminase changes were modest with AST increases generally higher than ALT.
• Our model identified those with COVID-19 from demographics and liver tests alone.
ALT | alanine aminotransferase |
AST | aspartate aminotransferase |
AUC | area under the curve |
CIHI | Canadian Institute for Health Information |
COVID-19 | coronavirus disease |
CSD | census subdivision |
DAD | discharge abstract data |
INR | international normalized ratio |
OHDP | Ontario Health Data Platform |
OHIP | Ontario Health Insurance Program |
OLIS | Ontario Laboratory Information System |
OLIS-C19 | Ontario Laboratories Information System – COVID-19 subset |
RIO | rurality index Ontario |
RPDB | the Ministry of Health Registered Person DataBase |
SaRS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
XGBoost | extreme gradient boost |
Starting in late 2019, a novel viral illness due to SARS-CoV-2 virus infection, known as COVID-19, circulated the globe widely, resulting in a pandemic leading to substantial morbidity and mortality.1,2 While much work has focused on how SARS-CoV-2 infection effects the respiratory system,3 it also effects many other organ systems, including the liver.4 Testing and monitoring for the SARS-CoV-2 at a population level is a major health concern. Improved understanding of population-level biomarkers for this infection could improve monitoring strategies and enable early identification of outbreaks.
Monitoring of SARS-CoV-2 transmission has been a major public health concern. Initially monitoring relied primarily on patient respiratory swabs, but population level wastewater monitoring has also been informative. Respiratory swab-based testing is not sustainable at a whole population-level.5 Additionally, wastewater monitoring lacks specificity and is a lagging indicator of community-level transmission.6 Further bellwether signals of increased transmission are needed.
Liver bloodwork abnormalities are independently significantly predictive of COVID-19 diagnosis with high accuracy.4 Abnormalities in alanine aminotransferase (ALT), aspartate aminotransferase (AST) and albumin are associated with COVID-19.4 Additionally, early reports during the pandemic identified significant abnormalities in ALT and albumin in individuals with confirmed COVID-19 compared to those with suspected COVID-19, and there was a significant correlation between AST alone and confirmed COVID-19.7 In the largest previous cohort study of liver bloodwork metrics, consisting of 417 individuals, more than 75% of individuals with COVID-19 had liver injury.8
Population level bloodwork anomalies could potentially act as an additional monitoring tool for infectious disease transmission and outbreak. During the COVID-19 pandemic, it was recognized that SARS-CoV-2 caused a hepatitis, consistent with other similar viral infections, but only in some individuals, especially those with more severe disease in other systems.9 Although liver bloodwork abnormalities in this scenario are non-specific, they are widely tested quantitative metrics, especially in acutely ill individuals with non-specific symptoms. The extensive use of this testing means it may be possible to monitor liver bloodwork at a population level to detect increased viral transmission. This could provide an ancillary measure for public health units to detect early viral outbreaks and increased transmissible illness.
The COVID-19 pandemic provides a real-world experiment scenario where there was extensive testing and reporting for SARS-CoV-2. In the Canadian province of Ontario, a centralized single-payer (government) health care system has allowed for population-wide data collection of both SARS-CoV-2 infections and liver bloodwork down to an individual level. Almost 15 million individuals live in Ontario (Ontario Data Catalogue, 2021). Thus, the relationship between a circulating infectious illness (COVID-19) and population level liver bloodwork can be explored.
Additionally, there has been recent focus on viral-induced hepatitis specifically in children, due to clusters of pediatric idiopathic acute hepatitis across the globe. These clusters have been linked to adeno-associated virus 2 (AAV2) potentially in combination with concurrent infections with other viruses, including SARS-CoV-2.10 This phenomenon appeared limited to children, mostly those less than 10 years old. Thus, we examined a pediatric subcohort in the current study.
We hypothesized that there is an epidemiologic link between abnormal liver bloodwork in relation to increased regional incidence of COVID-19. This study leverages the massive Ontario health administrative datasets, to examine for liver-related bloodwork changes within the 28 days after individuals have a positive COVID-19 test. Since there are multiple liver-related bloodwork tests available, and the same illness appears to cause varying changes in these tests, we used machine learning techniques to identify patterns in this dataset that would not be otherwise discernable.
This study was approved by the Queen’s University Health Sciences Research Ethics Board (PAED-542-22) and the Ontario Ministry of Health for access on the Ontario Health Data Platform (OHDP). Consent to participate was not applicable, as administrative databases were used for data collection. The data for this study derives from OHDP, a high-performance computing environment that linked large health datasets from Ontario, Canada, serving a population of approximately 14.8 million (Ontario Data Catalogue, 2021). The data analysis plan was not preregistered.
The dataset for the present study was created by extracting variables from the following databases within the OHDP: Ontario Health Insurance Program (OHIP) for diagnostic codes, the Canadian Institute for Health Information (CIHI) discharge abstract data (DAD) for hospitalized patient information, the Ontario Laboratory Information System (OLIS) for liver related laboratory values, and the Ontario Laboratories Information System – COVID-19 subset (OLIS-C19) for any positive test for any SARS-CoV-2 variant. Further individual demographic information (year of birth, sex, geographic location information) was obtained through the Ministry of Health Registered Person DataBase (RPDB). All direct individual identifiers were removed from the data prior to analysis. Supplemental materials are available via Zenodo at doi.org/10.5281/zenodo.18236199.11
The study period was from January 4, 2020 to September 2, 2022 (for COVID-19 tests) with follow up to 28 days after the last index date for labs. Inclusion were: any individuals over one year old (with no upper age limit) at the time of their COVID-19 test in the Ontario health administrative dataset (all were polymerase chain reaction tests for any variant), who have at least one positive COVID-19 test within the study period and at least one subsequent liver-related blood test in the 28 days following the positive test. Liver-related blood tests used in this study were based on clinical guideline recommendations for investigating liver disease: ALT, AST, international normalized ratio (INR), conjugated bilirubin, total bilirubin, and albumin.12
Data was extracted for cases at the time of their first positive COVID-19 test, regardless of if they had or had not had previous negative tests. If there were more than one of any given liver test in the 28 day period, then the most severely abnormal value was taken in the direction of the test that would indicate liver injury (that is, increased ALT/AST/INR/bilirubin levels, and decreased albumin levels). The values used to define normal ranges across sex and age groups was used for each test is found in Supplemental Table S1. Individuals excluded were those who: had any pre-existing billing diagnosis of hepatitis or cirrhosis (OHIP diagnostic codes 70 and 571, respectively) in DAD and OHIP (prior to their first COVID-19 test), incomplete demographic data, and individuals less than 1 year old.
Matched controls were individuals with their first negative COVID-19 test within the study period and at least one subsequent liver-related blood test (as defined above) in the 28 days following the negative test and no further COVID-19 test within the subsequent 28 days. These individuals otherwise met the inclusion criteria. COVID-19 positive cases were then each matched by age (in years) and sex with two control individuals, with the same exclusion criteria. Controls were always unique individuals from cases, by their unique identifier. Thus, those who were controls that at a later date were otherwise eligible to become cases (by later having a positive COVID-19 test and otherwise met the inclusion criteria) were excluded. The data extraction process is summarized in Figure 1.
All analysis was performed within the OHDP environment, using R (version 4.1.3) and python (v 3.8.5). Comparisons of COVID-19 positive versus negative individuals used the Person’s chi-squared test with Yates’ continuity correction. Due to the non-parametric distribution of all the liver blood tests (most were heavily right skewed), we used the Mann-Whitney U test to compare each blood test for between cases and controls. Associations were performed using logistic regression function in the base R stats package. Statistical significance was considered a p-value less than 0.05.
For machine learning, the sklearn, LogisticRegression, RandomForestClassifier, XGBoost, and TensorFlow python packages were used. The dataset included all liver blood tests (except conjugated bilirubin, since it was strongly correlated with total bilirubin), age (in years), and sex. For the complete data-only models Rurality Index for Ontario measures were also included for each individual based on their associated census subdivision (CSD) number recorded in RPDB.13 The data was divided 80:20 into training and testing sets. Hyperparameters were tuned using a focused iterative search strategy. We implemented a logistic regression model configured with an L1 penalty, a regularization strength of 0.1, and a liblinear solver. We implemented a random forest classifier configured with 100 estimators, as well as an XGBoost classifier that was configured with a learning rate of 0.1, a max depth of 5, and a logloss evaluation metric. We also developed a fully connected neural network consisting of five hidden layers, each with thirty neurons, that uses the ReLU activation function connected to a single neuron output layer with sigmoid activation for binary classification. The networks was configured with an Adam optimizer and a binary cross-entropy loss function. The network was trained for 50 epochs with a batch size of 100. The testing set was only evaluated once, after all hyperparameters were set and the model fully developed on the training set.
101,574 cases were age and sex matched to 200,709 matched controls met the inclusion/exclusion criteria ( Figure 1). Of the cases, 49% (49,299) had at least one abnormal liver blood test in the 28 days following their positive COVID-19 test. Of the controls, 41% (82,495) had at least one abnormal liver blood test in the 28 days following their negative COVID-19 test. Individual demographics and blood test statistics are summarized in Table 1.
Multiple differences were found between individual liver test values when comparing individuals with a positive COVID-19 test (cases) to those with a negative COVID-19 test (controls). The rate of abnormal tests, when accounting for differences in normal ranges by age, was statistically significant for all six liver blood tests between cases and controls ( Figure 2A, with summary statistics in Supplemental Table S2).
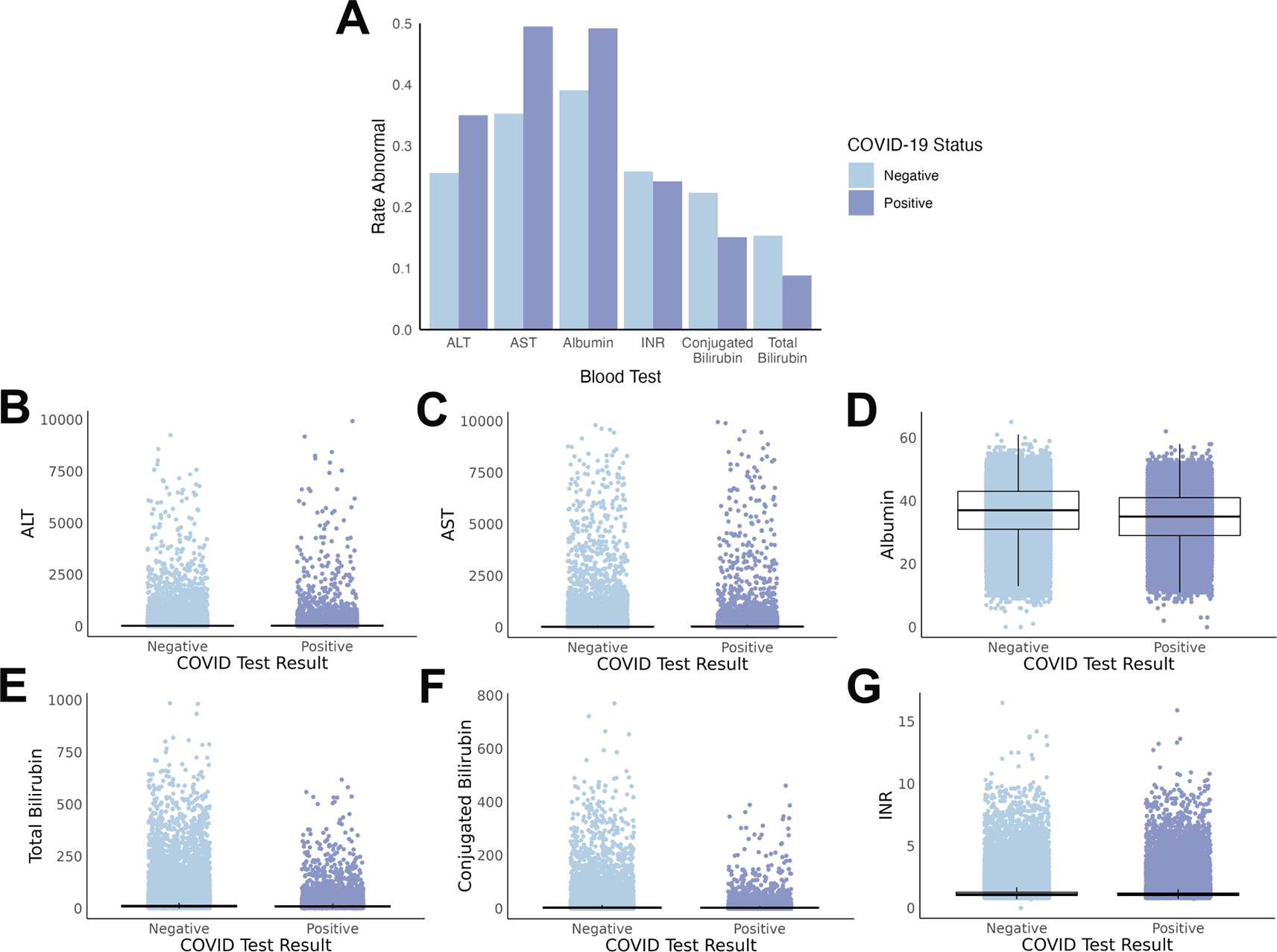
(A) Rate of abnormal test results differ by COVID-19 status, with overall increases with COVID-19 for ALT, AST, and albumin, and decreases with COVID-19 for INR, conjugated bilirubin, and total bilirubin. (B-G) Scatterplots demonstrate the substantial overlap between the absolute values of each liver test at a population level. Boxes represent the interquartile range (IQR) with the median value inside the box and the vertical lines represent the smallest and largest data points within 1.5 times the respective IQR. All absolute value tests, except INR, were statistically significant between cases and controls.
The degree of alteration to liver blood tests is also important, not simply the abnormal/normal dichotomy. Thus, we also compared the absolute values of the individual tests between cases and controls ( Figure 2B-G). In this comparison, five out of the six test absolute values were significantly different, with INR the exception (summary statistics in Supplemental Table S2).
Since the study duration extended over 28 days following the COVID-19 test, we examined the frequency at which each test was performed in each group over this time. It would be important to identify if certain tests were more commonly done later than others as these would be theoretically more likely to be normal and thus could bias the machine learning models. However, the trends of dates of each test were similar with most of the testing being done within 3 days of the individual’s COVID-19 test and then rapidly trending down to a low rate of further testing after that for all tests (Supplemental Figure S1). Notably, there are peaks for the frequency of each test at 7-day intervals, likely representing practitioners monitoring these tests weekly.
To identify variables with high likelihood to change together, we performed a correlation analysis of each permutation of pairs of liver tests (Supplemental Figure S2). Of the 15 pair-wise correlations, only ALT/AST and total bilirubin/conjugated bilirubin were statistically correlated (r-squared values of 0.56 and 0.88, respectively).
We initially compared four machine learning models with all matched individuals that met the inclusion criteria for the study. We chose to exclude conjugated bilirubin since it was highly correlated with total bilirubin, and total bilirubin was more commonly tested. We chose not to dimension reduce these two parameters into one since the explainability of the model could be important and so we kept the clinically relevant value. ALT and AST were not dimension reduced despite a modest correlation, since these parameters are both clinically important and their relative contributions to the models would be important information for determining the clinical value of these tests in suspected COVID-19 cases.
We compared logistic regression, random forest, extreme gradient boost (XGBoost), and multilayer perceptron neutral network models. The outcome was COVID-19 status (positive/negative). After hyperparameter tuning on the training set, each model was used to evaluate the test set, and the optimal model was XGBoost with an area under the curve (AUC) in the receiver operator characteristic curve of 0.71 ( Figure 3A). Notably, age was the feature with the highest relative contribution to the model (F score = 469), with four of the five blood tests contributing relatively highly to the model (F scores range of 366 to 425). INR contributed modestly with an F score of 36 and sex contributed minimally with an F score of 55 ( Figure 3B).
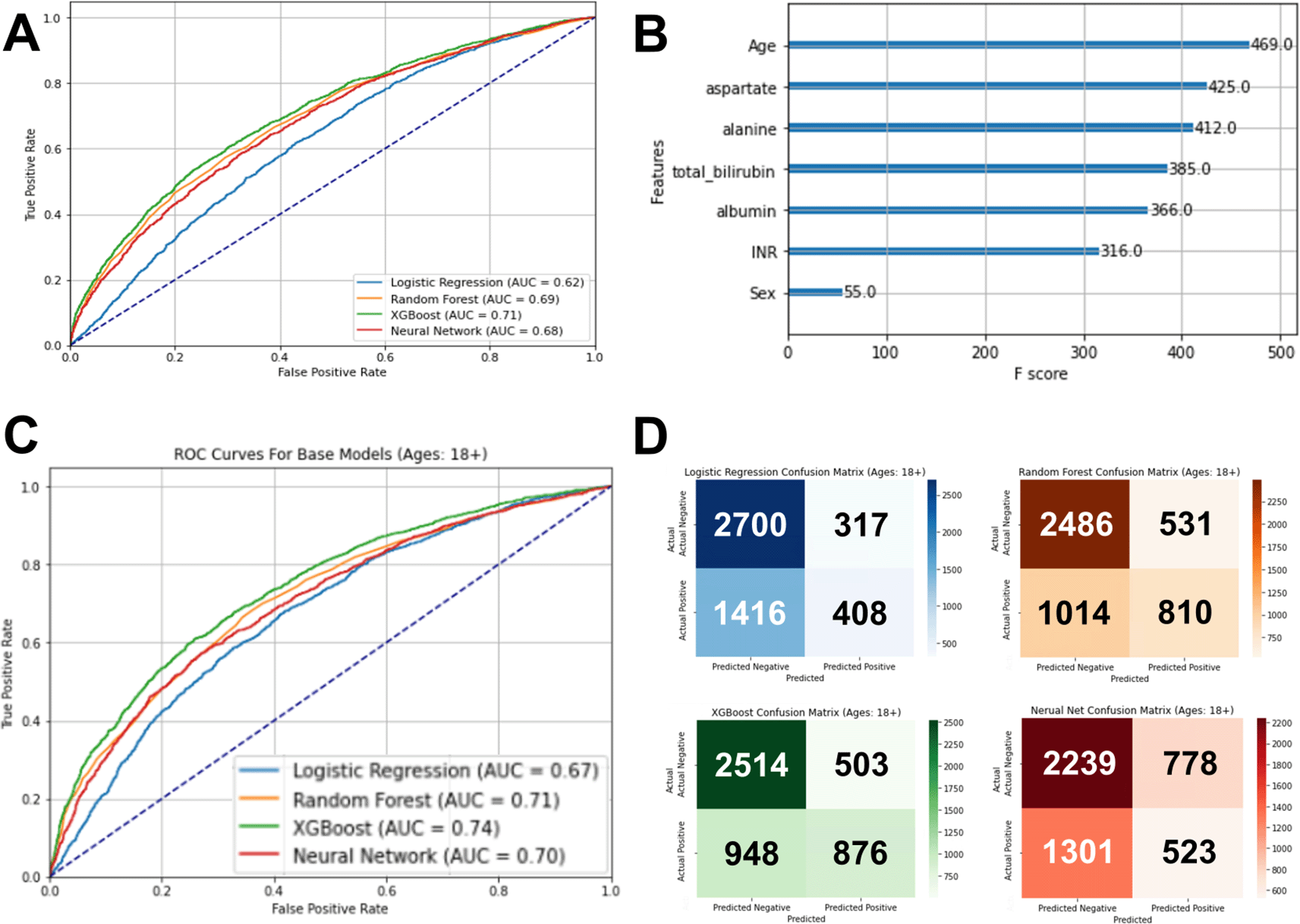
(A) The optimal model using XGBoost had an area under the receiver operator curve of 0.71. (B) In the optimal model, age was the feature with the highest relative importance to this model, followed by the five liver tests. (C) The XGBoost model for the adult patient subcohort test set was the optimal model, having an area under the receiver operator curve of 0.74. (D) Confusion matrixes of each machine learning model for the adult patient subcohort test set showing a balance of sensitivity and specificity for the XGBoost model, compared to the other models.
We suspected that the sparse nature of the data (as seen in Table 1) was limiting the models’ ability to identify COVID-19 cases, so we then limited the cohort to only individuals with all five of the blood tests available (excluding conjugated bilirubin). We also added the rurality index Ontario (RIO) as a variable. Notably, with this data subset, there were no missing values and included 24,676 individuals ( Table 2).
Since the impact of liver disease can differ by age group, and the general models had age having a relatively strong impact on the models, we also performed subgroup analysis on the larger cohort limiting to only individuals less than 18 years old, only those 19-64 years old, and only those greater than 64 years old (and permutations of pairs of these three age groups). The AUC improved most with those less than 18 years old excluded ( Table 3). Again, the optimal performing model was XGBoost, with an increase in the AUC to 0.74 on the test set ( Figure 3C and D).
The present study used a large health administrative dataset to demonstrate that abnormal liver bloodwork is common in SARS-CoV-2 infection. The pattern of liver test abnormalities showed that a subset of individuals with COVID-19 had elevated ALT, AST, and lower albumin compared to the control population. The transaminase changes were modest with AST increases generally higher than ALT. A machine learning model was able to distinguish individuals with COVID-19 from control individuals using demographics and liver blood tests alone. The ability of the model to classify individuals correctly was improved when the dataset was limited to only adult individuals.
The impact of COVID-19 infection on the liver is unclear at present, likely due to individual heterogeneity. Some studies demonstrate relatively minimal changes to ALT and AST in the large majority individuals with COVID-19 infection, with most having these enzymes in the normal range.14 Other studies show a substantial number (14-53%) of individuals with COVID-19 also having abnormal ALT and AST, and also these changes were associated increased mortality.15 Notably, these studies analyzed cohorts that were orders of magnitude smaller than our study.
It seems likely that liver-related bloodwork changes in COVID-19 are a direct result of a viral hepatitis but this is not entirely clear at present.16–18 COVID-19 infection is unique in many important ways from other viruses that cause hepatitis, in particular, it is relatively novel, and highly associated with vascular injury, hypoxic injury, and cytokine storm in some individuals.19 Thus, liver-related bloodwork changes in COVID-19 are important to better understand as they specific patterns of changes could inform clinical decision making. Luca et al. showed AST elevations were higher than ALT elevations in COVID-19 infection, at a population level, which is the opposite of classic dogma around viral hepatitis patterns.15,20 Our study shows the same broad trend of AST elevation higher than ALT, although the present study is large enough to suggest that these ALT and AST differences are clinically relatively similar, as seen in Table 1.
The statistically significant differences in the ALT and AST data demonstrates greater differences in the medians than the means between the case and control cohorts, indicating that the difference is driven at least in part by outliers. Clinically, this may indicate that individuals with very high ALT and AST elevations are more likely to have COVID-19 infection. Additionally, the other liver tests included in our study (albumin, bilirubin, and INR) despite being statistically different, had only clinically modest differences in cases comparted to controls.
Unique to our study, we observed that cases had lower rurality index than controls in Table 2. This index is based on a weighted scoring combining population density with distance to basic and advanced health care facilities. Access to testing could have influenced this difference, but it may also be that the rate of COVID-19 was higher in more densely populated areas causing there to be more cases to come from more urban settings.
This study is limited in that it only had access to health administrative-level data, which is limited to demographic and objective test-related data, but not record level details about the individual’s overall health. However, this study does have a balancing strength of having a very large cohort available for analysis compared to previous similar studies.14,15 It is possible that more common liver related illnesses are present but not previously identified by diagnostic codes in our study (such as metabolic disease-related conditions and alcohol-associated liver disease).21 However, it is likely that in a cohort this large, individuals with these diagnoses would likely be present in both the case and control cohorts relatively in similar rates. Additionally, this study is limited to individuals who recently had a COVID-19 test so there is likely some confounding bias from other infections in the control group. Nevertheless, this would likely only moderate the statistical differences we did find between the case and control groups.
Another important limitation is that during the study period, there were shifts in the severity and clinical impact of SARS CoV-2, namely through evolution of variants and the increasing availability of vaccination. The study dataset did not include variant or vaccination information so, unfortunately, we are unable to separate out these important variables. Future studies that explore these important differences would be of further value to understanding the role of variants and vaccines that could assist in preparation for future viral pandemics. Race and ethnicity are important demographic factors for COVID-19 and for liver disease. Unfortunately, race-ethnicity information was not available for our cohort, as it was not included in the study design.
The machine learning models in this study were not expected to be able to diagnose COVID-19 from liver-related bloodwork, given the number of confounding conditions present in the general population that can cause alterations in liver-related bloodwork. Instead, these exploratory models were able to assist with showing unique patterns in liver-related bloodwork between those with and without COVID-19. These unique features could help clinicians with understanding the typical liver-related bloodwork patterns in COVID-19 patients, potentially helping where the diagnosis may not be clear or there may be multiple comorbidities. Additionally, it is useful for clinicians to know that our study suggests there is no single clear pattern of liver-related bloodwork changes that are pathognomonic for COVID-19.
In conclusion, this large cohort study has demonstrated decisive differences and patterns in liver blood tests in individuals with COVID-19 infection. These differences support previous findings from smaller studies. These liver blood test differences alone can be linked to COVID-19 infection with modest confidence using machine learning.
This study was approved by the Queen’s University Health Sciences Research Ethics Board (PAED-542-22) and the Ontario Ministry of Health for access on the Ontario Health Data Platform (OHDP). Consent to participate declaration: not applicable, as administrative databases were used for data collection.
This study used de-identified personal health information. Due to the sensitive nature of the data, raw data remains confidential and cannot be shared. The dataset supporting the conclusions of this article is restricted, as it includes personal health data, and is available through ICES, with further information available at: https://datadictionary.ices.on.ca/Applications/DataDictionary/Usage.aspx.
[Supplemental Materials]
Supplementary material intended for this study is added to the following link. Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
Mulder, DJ; 2026; Supplemental Materials for SARS-CoV-2 Infection Impacts Liver-Related Blood Tests Following Infection: A Health Administrative Data Study; Zenodo; doi.org/10.5281/zenodo.18236199. (11)
Parts of this material are based on data and information compiled and provided by Ontario Ministry of Health (through Ontario Health Data Platform). However, the analyses, conclusions, opinions and statements expressed herein are those of the author, and not necessarily those of the Ministry. Parts of this material are based on data and information compiled and provided by the Canadian Institute of Health Information (CIHI). However, the analyses, conclusions, opinions and statements expressed herein are those of the author, and not necessarily those of CIHI. This study was supported by the ICES, which is funded by an annual grant from the Ontario MOH. The opinions, results and conclusions reported in this paper are those of the authors and are independent from the funding sources. No endorsement by ICES or the Ontario MOH is intended or should be inferred. Parts of this material are based on data and information provided by Ontario Health (OH). The opinions, reviews, view and conclusions reported in this publication are those of the authors and do not necessarily reflect those of OH. No endorsement by OH is intended or should be inferred.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)