Keywords
Staphylococcus, eye infections, PCR, coagulase-negative staphylococci, antibiotic resistance, mecA, icaA, tetk
This article is included in the Fallujah Multidisciplinary Science and Innovation gateway.
The most typical bacterial species that may be associated with infections of the eye are Staphylococcus because they normally reside on the skin and eyelid margins; therefore, an easy transmission to the eye can take place whenever there is a lesion or abrasion. Apart from this, these bacteria have several virulence factors among which include adhesin and biofilm forming capacity which enhance their resistance against treatment. In addition, mecA gene carries resistance determinant against methicillin as well as many other β-lactam antibiotics.
The current study was designed to characterize the factors and molecular mechanisms that facilitate pathogenicity and therapeutic resistance in these bacteria. A total of one hundred samples were obtained from patients who had visited Samarra General Hospital during the period extending from October 2024 to December 2024. As per the results of culture, all isolates were Gram-positive, and forty-nine belonged to the genus Staphylococcus. Amongst these, Staphylococcus aureus was the major species followed by Staphylococcus epidermidis and some other isolates.
All isolates showed 100% resistance to Methicillin. They were sensitive to Vancomycin. Other antibiotics expressed variable sensitivity patterns among the isolates. The five isolates that expressed the highest levels of antibiotic resistance were selected, and one was excluded during analysis. All the remaining isolates, being coagulase-negative staphylococci, were tested for mecA, icaA, and tetK genes.
The molecular results showed that all four tested isolates carried the mecA gene (100%), and only one isolate carried the icaA and tetK genes, hence its high level of antibiotic resistance.
Staphylococcus, eye infections, PCR, coagulase-negative staphylococci, antibiotic resistance, mecA, icaA, tetk
The human eye is the most important sensory organ involved in the act of vision. It occupies an imperative place in perception and recognition of objects, forms, and environment. Several related parts of the eye work in collaboration with some other organs, especially the brain and nerves, to bring about clear and complete vision.1
The eyes have much defense mechanism since they are constantly exposed. Tears and their secretion together with the movement of eyelids help remove pathogenic particles. Secretory enzymes also function to enhance protection, meanwhile antibodies give more other forms of support. Normal microbial flora of the conjunctiva is also very useful in the defense of the eye.2
These defense mechanisms are not foolproof; various microorganisms still manage to invade the eye and cause infection. The infections could be of bacterial, viral, or fungal origin. However, since bacteria predominate among all ocular infections due to their virulence factors and because of host immunity weakness as well as age and poor personal hygiene. The infections could be simple conjunctivitis or severe keratitis or endophthalmitis which may sometimes lead to vision loss.3
Gram-positive bacteria have been regarded as the main etiological agents of infections of the eyes, and in these, the genus Staphylococcus is one of the most prevalent. Several studies have described Staphylococcus aureus as the leading etiological agent for eye infections due to its multiple virulence factors which include collagen-binding proteins as well as production of an adhesive mucous layer that facilitates attachment to host tissues.4
Methicillin-resistant Staphylococcus aureus strains have now emerged. This poses a serious clinical challenge because the resistance is derived from plasmids and resistance genes that are transferable, together with changes in the drug target site. Indiscriminate as well as excessive usage of antibiotics for treatment of bacterial and viral infections or for prophylaxis has led to increased rates of antibiotic resistance globally.5
Genes are basic building blocks of heredity in all living things coding information for the structure and function of biology. They usually locate in deoxyribonucleic acid (DNA), except seldom cases where they find in ribonucleic acid (RNA).6
Because the Staphylococcus species are important agents of ocular infections that can sometimes lead to serious complications and since antibiotic-resistant strains keep increasing, it has become imperative to study their molecular characteristics. Therefore, this study was designed to isolate and characterize at the molecular level Staphylococcus species according to different levels of resistance they bear against antibiotics.
One hundred samples were taken from patients with eye infections at Samarra General Hospital. Samples were collected from the conjunctiva using a sterile cotton swab under the supervision of an expert physician. The timeframe was from October 2024 to December 2024. All samples were directly transferred after collection to the research laboratory at the University of Samarra for processing.
Each swab sample was inoculated into 5 mL of Nutrient broth and kept at 37°C for 24 hours. After that, subcultures were done on selective media which included mannitol salt agar and blood agar then these were incubated aerobically at 37°C for another 24 hr. The obtained bacterial isolates were purified and Gram-stained to give a preliminary identification result. Colony shape, size, color, pigmentation as well as microscopic appearance, and cellular arrangement were noted.7,8 Catalase, oxidase, urease plus coagulase biochemical tests validated the bacterial species. The isolates were finally confirmed by the Vitek2 Compact System.
The samples were taken from participants who signed informed consent, following the ethical guidelines of the University of Samarra. The test for antibiotic susceptibility was carried out by the Kirby–Bauer disk diffusion method based on standard recommendations as described by the WHO. Ten antibiotic disks used included Ofloxacin (OFX, 30 μg), Levofloxacin (LEV, 5 μg), Ciprofloxacin (CIP, 5 μg), Gentamicin (CN, 10 μg), Neomycin (N, 30 μg), Tetracycline (TE, 30 μg), Vancomycin (VA, 30μg), Chloramphenicol (C, 30 μg), Trimethoprim (TM, 5 μg) and Methicillin (MET, 5μg).
The bacterial inoculum was surface spread onto Mueller–Hinton agar plates, and the antibiotic disks were applied to the surface. These were incubated at 37°C for 24 hours. After incubation, measurements of each diameter of inhibition zone were read and interpreted in accordance with international reference standards.9
DNA extraction followed the ABIOpure protocol in this manner: Centrifuge an overnight culture to pellet the cells and discard the supernatant. Add lysis buffers and enzymes specific for wall degradation and protein digestion, then wash and spin several times to purify DNA up to high purity nucleic acid that is finally eluted and stored at –20°C for further applications.
The Polymerase Chain Reaction (PCR) was then performed to amplify the target genes mecA, icaA, and tetK using gene-specific primers, as listed in Table 1. These primers were supplied by Macrogen in a lyophilized form. Each PCR mixture had a total volume of 20 μL, consisting of 10 μL of 2× Master Mix (containing Taq DNA polymerase, deoxynucleotide triphosphates (dNTPs), and magnesium ions), 1 μL each of the forward and reverse primers for each gene, 2 μL of template DNA extracted from bacterial isolates, and 6 μL of nuclease-free water to complete the total volume.
| Primer name | Seq. | Annealing temp. (°C) | Product size (bp) | Ref. |
|---|---|---|---|---|
| mecA-F | AAAATCGATAAAGGTTGGC | 51°C | 532 | 10 |
| mecA-R | AGTTCTGCAGTACCGGATTTGC | |||
| icaA-F | TCTCTTGCAGGAGCAATCAA | 50°C | 188 | 11 |
| icaA-R | TCAGGCACTAACATCCAGCA | |||
| tetK-F | GTAGCGACAATAGGTAATAGT | 54°C | 360 | 12 |
| tetK-R | GTAGTGACAATAAACCTCCTA |
Amplification was carried out using a Thermal Cycler according to the temperature program described in Table 2. The program began with an initial denaturation at 95°C for 5 minutes, followed by 30 amplification cycles, each consisting of denaturation at 95°C for 30 seconds, annealing for 30 seconds at gene-specific temperatures (50°C for icaA, 51°C for mecA, and 54°C for tetK), and extension at 72°C for 30 seconds. After cycling, a final extension was performed at 72°C for 7 minutes to ensure complete synthesis of all amplicons, and the reaction products were maintained at 10°C until analysis.
| Steps | °C | m:s | Cycle | |
|---|---|---|---|---|
| Initial Denaturation | 95 | 05:00 | 1 | |
| Denaturation | 95 | 00:30 | 30 | |
| Annealing | mecA | 51 | 00:30 | |
| icaA | 50 | 00:30 | ||
| tetK | 54 | 00:30 | ||
| Extension | 72 | 00:30 | ||
| Final extension | 72 | 07:00 | 1 | |
Verification of PCR products was conducted by agarose gel electrophoresis using a 1.5% agarose gel prepared in Tris–Borate–EDTA (TBE) buffer. The amplified DNA bands were stained with ethidium bromide and visualized under ultraviolet (UV) illumination to confirm the expected amplicon sizes by comparison with molecular weight markers.
A total of 100 samples were collected from patients suffering from eye infections. After culturing the samples obtained using sterile cotton swabs on various culture media, the results revealed that 73 samples (73%) showed positive bacterial growth, whereas 27 samples (27%) exhibited no bacterial growth, as illustrated in Figure 1.
The isolates were identified according to reference Prescott’s Microbiology based on their morphological characteristics, colony shape, and color, in addition to their ability to retain the Gram stain. The identification of the isolates was further confirmed using the Vitek2 Compact System. The results showed that all the growing isolates were Gram-positive , i.e., 73 isolates were positive for Gram stain. The results also revealed that 49 isolates belonged to the genus Staphylococcus, while the remaining isolates were excluded.
Staphylococcus aureus represented the highest proportion among the Gram-positive isolates, with a growth rate of 38% (19 isolates). Regarding coagulase-negative staphylococci (CONS), their isolates were predominant compared to coagulase-positive staphylococci, with 30 isolates, representing 61.2% of the total Gram-positive isolates.
These identified isolates belonged to five bacterial species Staphylococcus epidermidis, which accounted for 16.3% (8 isolates out of 49), followed by Staphylococcus hominis, Staphylococcus lugdunensis, and Staphylococcus warneri, each with 12.2% (6 isolates for each species). Four isolates of Staphylococcus sciuri were also obtained, representing 8.1%, as shown in Table 3.
Antibiotic susceptibility testing has revealed variation in the response of studied isolates to tested antibiotics. Results obtained indicated that all the isolates were 100% sensitive to Vancomycin, meanwhile all the isolates dwelled resistance against Methicillin. Sensitivity to other antibiotics elicited varied responses which amounted to a resistance percentage of 20.4% from the isolates for each of Gentamycin, Levofloxacin, and Ofloxacin.
Regarding the antibiotics Tetracycline, Trimethoprim, Ciprofloxacin, Chloramphenicol, and Neomycin, the bacterial resistance rates were 36.7%, 34.6%, 22.4%, 14.2%, and 6.1%, respectively, as shown in Table 4.
Five Staphylococcus bacterial isolates were selected for the detection of mecA, icaA, and tetK genes. They carry sequence numbers (37, 47, 49, 87, and 93). These isolates expressed the highest level of resistance against antibiotics.
Three of the isolates belonged to Staphylococcus epidermidis with sequence numbers 49, 87, and 93, while one isolate with sequence number 47 belonged to Staphylococcus hominis. The isolate carrying sequence number 37 was excluded because it was invalid.
The genes were analyzed using the specific primers for each gene and the PCR procedure described previously. The results of agarose gel electrophoresis showed that all isolates contained the mecA gene (100%), with a band size of 532 bp, which corresponded to the expected product size, as shown in Figure 2.
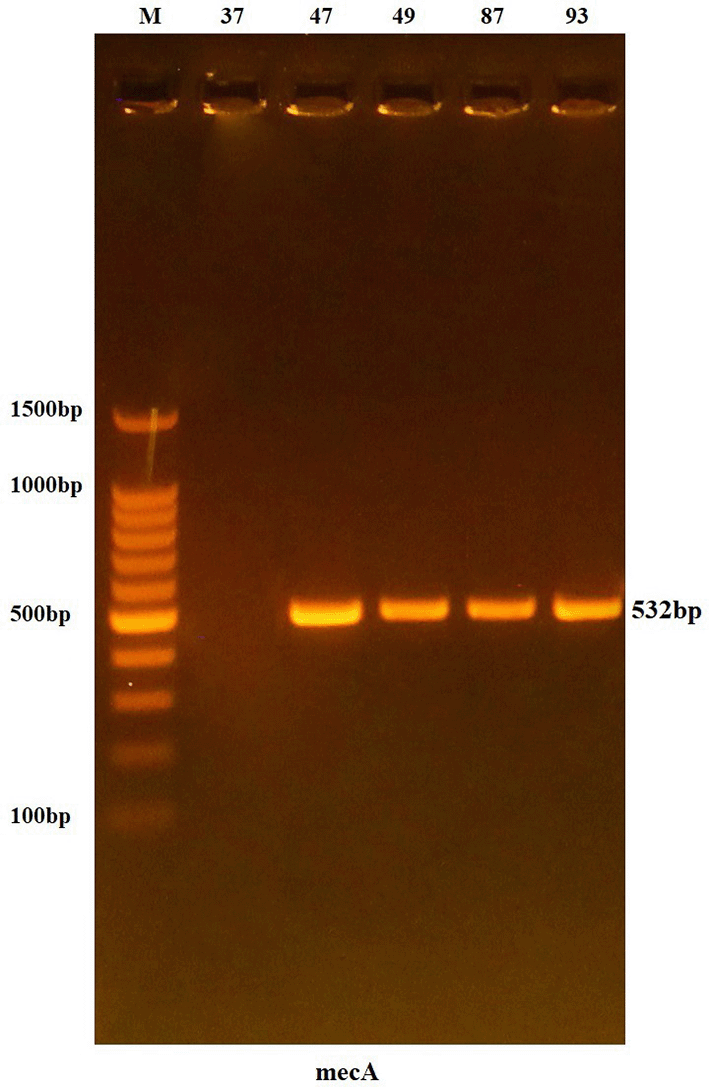
Lanes 37-93 resemble 532 bp PCR products.
As for the icaA and tetK genes, both were present in only one isolate, which was the isolate carrying sequence number 93, as shown in Figures 3 and 4.
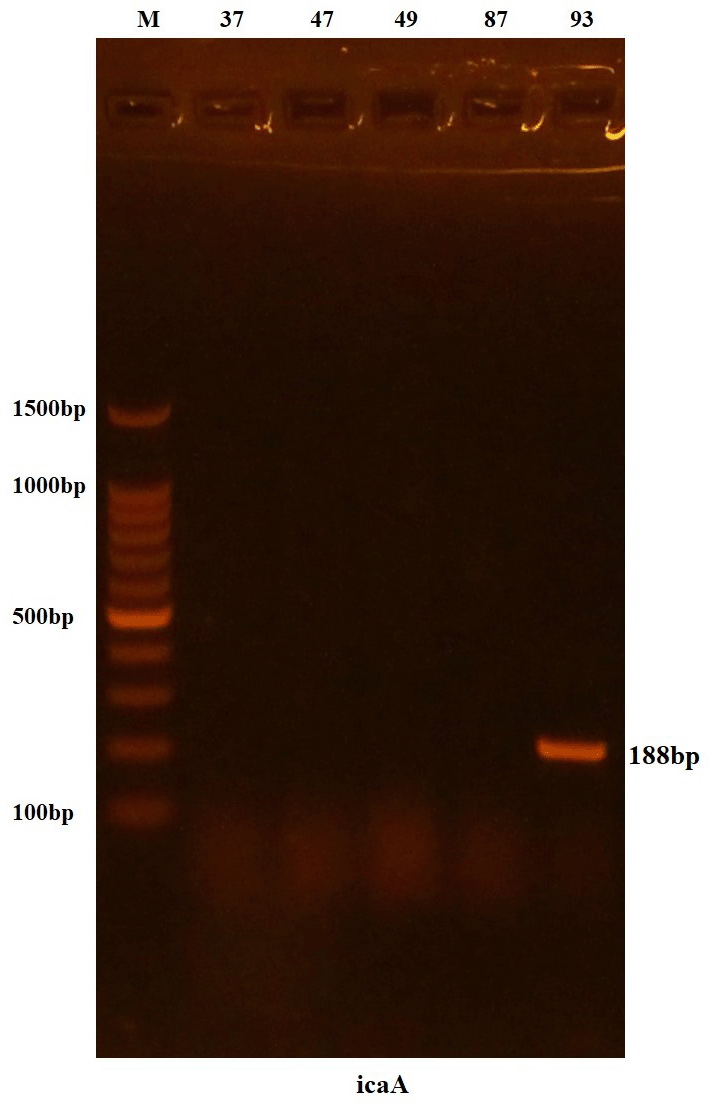
Lanes 37-93 resemble 188 bp PCR products.
A total of 100 samples were collected from patients suffering from eye infections who attended Samarra General Hospital. After culturing the samples, the results showed that 73 samples (73%) exhibited positive bacterial growth, while the remaining samples showed no growth. This may be attributed to the fact that some patients might have been taking antibiotics, which enhanced the body’s immune defense,13 or that the etiological agent of eye infection was viral or anaerobic bacteria.
After Gram staining, the results indicated that all growing isolates were Gram-positive , which is consistent with a study conducted in Samarra by Taha.14 This increase may be due to the chemical composition of the Gram-positive bacterial cell wall, which contains a high proportion of peptidoglycan, helping the bacteria to invade ocular tissues and inhibit the process of phagocytosis.15
49 isolates were identified as belonging the isolates were discarded. Results indicated that Staphylococcus aureus presented a higher growth rate representing 38% from the total isolates. This result goes with a study done in Erbil by Agha16 which showed that Staphylococcus aureus is among the common bacterial causative agents of ocular infections.
Coagulase-negative staph were the top bugs, with 30 isolates (61.2%) out of all Gram-positive bacteria. This number is near what Ali17 shared, noting that coagulase-negative staph made up 66% of Gram-positive isolates. The high count of CONS can be linked to their place as part of the normal skin germs.18
The most prevalent coagulase-negative species is Staphylococcus epidermidis at 16.3% (8 out of 49 isolates). This percentage tallies with a study carried out in the United States who confirmed S. epidermidis to be the most frequently isolated species among coagulase-negative staphylococcus from cases of ocular infections.19 Next were to the Staphylococcus genus and the rest of S. hominis, S. lugdunensis, and S. warneri which shared the slot equally at 12.2% (6 isolates each). Four isolates represented S. sciuri also (8.1%). This can probably be attributed to their high resistance rates against antibiotics since these bacteria are no longer viewed as mere contaminants but have evolved into significant players in hospital-acquired infections.20
All isolates were 100% sensitive to Vancomycin, this percentage is close t what was recorded by Al-Adeeb21 who found a sensitivity percentage of 91%, meanwhile all isolates were 100% resistant to Methicillin.
The resistance pattern to the other antibiotics used was as follows: Levofloxacin, Ofloxacin, and Gentamycin at 20.4% each. Resistance to Tetracycline was recorded at 36.7%, 34.6% for Trimethoprim, 22.4% for Ciprofloxacin, 14.2% for Chloramphenicol, and 6.1% for Neomycin.
The resistance rate to Chloramphenicol in this study was in agreement with the findings of Rumaid et al.,22 who reported a rate of 13%, although the results differed for the other antibiotics. This variation may be due to the genetic diversity among Staphylococcus isolates or to mutations that occur during bacterial culturing and transfer between different media.
The results of agarose gel electrophoresis showed that all isolates used in the study carried the mecA gene (100%), producing a band of 532 bp, identical to the expected size, as shown in Figure 2. This high percentage is consistent with the findings of Dhiaa Al-Din, 23 who confirmed the presence of the mecA gene in 95.4% of bacterial isolates causing ocular infections. Similarly, another study on CONS bacteria isolated from eye infections reported a high mecA gene prevalence of 71%.24
One out of four isolates was positive, i.e., 25% with a band size of 188 bp corresponding to the expected product. This goes hand in hand with a German study which confirmed that some S. epidermidis isolates do have the icaA gene responsible for biofilm formation while it was absent in S. hominis.25 It may be attributed to the ability of Staphylococcus spp. to form biofilm by alternative mechanisms or sequence differences in genes preventing recognition by primers.
The tetK gene was seen in only one isolate (25%) and produced a band size of 361 bp as shown in Figure 4. This is the same isolate that carried the icaA gene hence its high antibiotic resistance. Altayb et al.,26 noted that S. epidermidis usually carries multidrug resistance, mecA, tetK, and icaA genes. The combination increases the stability and persistence of clinical environments.
Only Gram-positive bacteria were isolated. The species with the highest rate of infection among Staphylococci was Staphylococcus aureus. All the isolates showed resistance to Methicillin but sensitivity to Vancomycin. This, therefore, supports the argument that eye infection coagulase-negative staphylococcal isolates may house multiple resistance genes thus emphasizing their role as real pathogenic agents and not as contaminants.
Other microbial agents composed of viruses, fungi, and anaerobic bacteria that invade the eye should be further studied. Additional molecular studies about Staphylococcus epidermidis in expressing its pathogenic and resistance mechanisms are recommended. It is also recommended to perform wider molecular investigations of the icaA and tetK genes to the extent of their distribution among ocular isolates.
The study was conducted in accordance with the Declaration of Helsinki. Ethical approval was obtained from the Scientific Research Ethics Committee of the College of Education, University of Samarra, Samarra, Iraq (Approval No. 306, 29 August 2024). Written informed consent was obtained from all participants.
Figshare. Antibiotic susceptibility and PCR results of Staphylococcus spp. Isolated from Eye infection.
https://doi.org/10.6084/m9.figshare.30815819
The project contains the following underlying data:
Table:
paper1: shows the results of the antibiotic susceptibility test.
paper 2: Genotyping Results.
paper 3: Vitek Test Results.
paper 4: Coagulase Test Results.
paper 5: DNA Extraction Results.
Figshare. Antibiotic susceptibility and PCR results of Staphylococcus spp. Isolated from Eye infection.
https://doi.org/10.6084/m9.figshare.30815819
A compressed file containing:
Figure 1: Ethical approval for conducting the research.
Figure 2: Informed consent from patients participating in the research.
The data underlying this study are available under the terms of the (CC BY 4.0). This allows for the sharing and adaptation of the data, provided that appropriate credit is given to the original authors.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)