Introduction
The dsRNA binding protein (dsRBP) PACT (referred to as RAX in the mouse) activates the dsRNA dependent protein kinase PKR in response to various stress signals1–5. This in turn results in repression of translation through phosphorylation of the eukaryotic translation initiation factor eIF2α6. Although the biochemical function of PACT as an activator of PKR is well established, the physiological role of this protein in vivo is unclear. Indeed, generation of mice lacking RAX revealed a role for this protein in craniofacial7,8 and anterior pituitary development9 that is not readily explained by current knowledge of its biochemical function. More recently, PACT has been identified as a component of the RNA silencing (siRNA/miRNA) pathway10,11, although a detailed understanding of its role in this pathway is limited. Thus, PACT functions in two independent pathways that govern post-transcriptional gene regulation and as such it is important to understand the overall physiological function of this protein. To this end, we investigated the functional orthology between PACT and the Drosophila melanogaster dsRBP R2D2.
RNA silencing through the micro-RNA (miRNA) and short-interfering RNA (siRNA) pathways regulates nearly every aspect of cellular biology (reviewed by van Kouwenhove et al.12,13). Both siRNA and miRNA pathways require cytoplasmic processing of precursor molecules (either dsRNA or ~70bp pre-miRNAs) to generate mature guide RNAs (reviewed by van Kouwenhove et al.12). In both cases, this cytoplasmic processing step is accomplished by the endonuclease Dicer in complex with one or more dsRNA binding proteins, which cleaves precursor RNA molecules into ~20–25bp dsRNA. One of the two strands of the Dicer-processed mature RNA molecule (known as the guide strand) is then loaded into the effector complex, the RNA-induced silencing complex (RISC) (reviewed by Czech and Hannon13). Although the precise composition of the RISC is unclear, various studies have demonstrated the presence of a mature guide RNA, Dicer, one or more accessory dsRBPs, and one of the four members of the Argonaute protein family (Ago1–4) which function as the effector enzymes in the terminal silencing step14.
The RNA silencing pathways of metazoans have largely been investigated in the model organisms Caenorhabditis elegans and Drosophila melanogaster. In both systems, it was discovered that the endonucleases required for cleavage of pri-miRNAs (Drosha) and pre-miRNAs/siRNA precursors (Dicer) form complexes with specific dsRBPs. These proteins, like PACT, are comprised of tandem dsRBDs with no other identifiable domains (Drosha complexes with Pasha/DGCR815, C. elegans Dicer binds RDE-416, D. melanogaster Dicer-1 binds Loquacious (LOQS)17 and D. melanogaster Dicer-2 binds R2D218). Furthermore, these dsRBPs are required for the processing functions of the complexes. In mammalian systems, processing of both siRNA and miRNA is performed by a common Dicer enzyme. Both PACT and a similar dsRBP Tat Trans-activation element RNA binding protein (TRBP) in human/Protamine-1 RNA binding protein (PRBP) in mouse have been demonstrated to bind Dicer through either their N-terminal dsRNA binding motifs or their C-terminal Merlin-Dicer-PACT Liaison (Medipal) domain (which contains the PKR activation domain of PACT)10,11,19. The binding of PACT, TRBP or both with Dicer enhances the ability of Dicer to process dsRNA into siRNAs in vitro10. These findings suggest a functional significance to the PACT/TRBP/Dicer interactions, although the mechanism by which this occurs is unclear. These reports suggest that in addition to the regulating PKR activation, PACT may affect post-transcriptional control via small RNA pathways.
In Drosophila, processing of small RNA precursors is accomplished by separate enzymes, Dicer-1 for miRNA and Dicer-2 for siRNA20. The dsRNA binding proteins LOQS17 and R2D218 bind to Dicer-1 and Dicer-2, respectively. LOQS and R2D2 sort precursor RNA molecules based on the presence or absence of mismatches in the stemloop of the hairpin or dsRNA precursor to the appropriate Dicer processing pathway23 and facilitate RISC loading24,25. LOQS encodes four isoforms; LOQS-PA and LOQS-PB contain three tandem dsRBDs whereas both LOQS-PC and LOQS-PD21,22 lack the third C-terminal dsRBD. R2D2 encodes a single isoform that is comprised of two dsRBDs18 (Figure 1). Within the dsRBDs of these proteins, there is a high degree of sequence similarity (71–78% similarity, see Table 1) between PACT and LOQS PA and PB (which have all three dsRBDs in common). This sequence similarity coupled with the tandem dsRBD architecture (see Figure 1 and Table 1) indicates potential functional orthology between PACT and LOQS. Although R2D2 is significantly less similar to PACT (46% sequence similarity within dsRBD2, with no detectable similarity within dsRBD1), it has nonetheless been demonstrated to bind Dicer-218 and possesses a tandem dsRBD architecture similar to PACT and LOQS and thus may also function orthologously to PACT/RAX.
Table 1. Amino acid sequence similarity between PACT and Drosophila dsRBPs.
Amino acid sequence identity and similarity (positives) between dsRNA binding domains of PACT and the Drosophila dsRNA binding proteins determined by BLAST (blastp) alignment; ~ indicates no detectable similarity. The three dsRBDs shown for LOQS correspond to the shared dsRBDs of PA and PB isoforms; the PC and PD isoforms share the first and second but lack the third dsRBD.
| | PACT dsRBD1 | PACT dsRBD2 | PACT dsRBD3 |
|---|
| | Identity | Similarity | Identity | Similarity | Identity | Similarity |
|---|
| LOQS | dsRBD1
dsRBD2
dsRBD3 | 55%
~
~ | 71%
~
~ | ~
62%
~ | ~
78%
~ | ~
~
57% | ~
~
76% |
| R2D2 | dsRBD1
dsRBD2 | ~
~ | ~
~ | ~
26% | ~
46% | ~
~ | ~
~ |
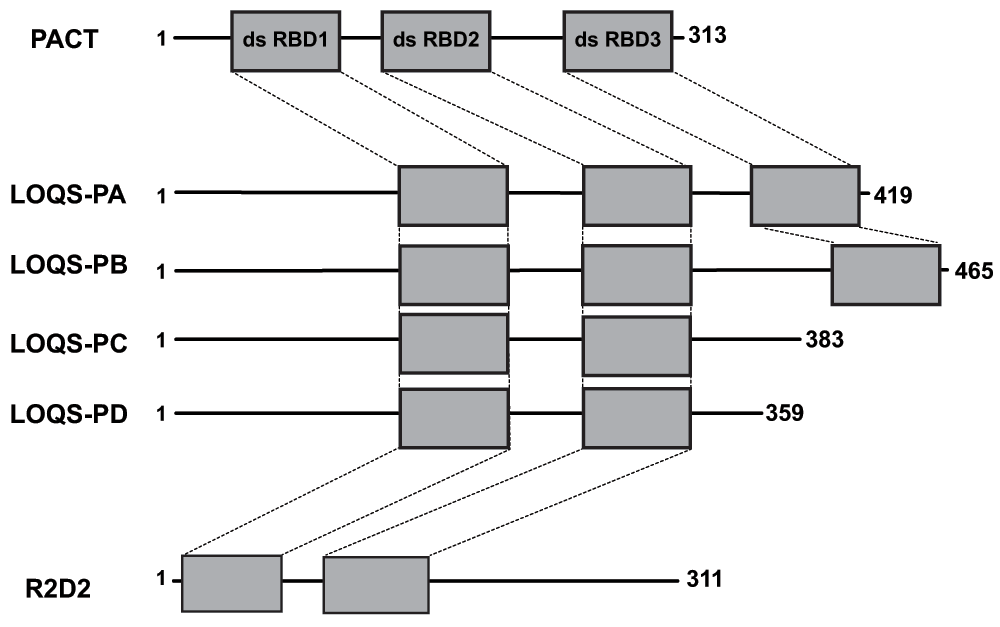
Figure 1. Schematic representation of PACT and Drosophila dsRBPs.
Schematic diagram of the PACT, LOQS and R2D2 proteins depicting the organization of their dsRBD domains. Dashed lines designate dsRBD1, 2 or 3.
Previous studies have produced mutant D. melanogaster lines lacking either of the dsRBPs that interact with Dicer-1 and Dicer-2. Flies deficient in all four isoforms of LOQS display developmental lethality, and do not survive past the pupal stage26. In contrast, flies homozygous mutant for a null allele of r2d2 are semi-lethal but the surviving flies have severe fertility defects27. The fertility phenotype results from defective oogenesis. Specifically, loss of R2D2 disrupts formation of the stalk cells, which normally separate individual follicles from each other28. This phenotype is enhanced by a dicer-1 mutant, indicating an unexpected interaction between R2D2 and Dicer-1 rather than Dicer-227.
We hypothesized that one or more dsRNA binding proteins with known function in D. melanogaster are functionally orthologous to PACT, and that identification of the functional ortholog(s) would provide insight into the small RNA processing function of PACT. We sought to identify proteins orthologous to PACT by transgenically expressing human PACT in the well-characterized Drosophila genetic model system. We report here our efforts to determine the degree to which human PACT is a functional ortholog of Drosophila R2D2.
Materials and methods
Cell lines, plasmids and antibodies
HeLa cells (ATCC CCL-2) were maintained in Dulbecco's modified Eagle's medium (DMEM) containing glucose (4.5g/L), penicillin (50U/ml), streptomycin (50µg/ml), L-glutamine (2mM) and sodium pyruvate (1mM) (Cleveland Clinic Lerner Research Institute Media Preparation Core) supplemented with 10% fetal bovine serum (Atlanta Biologicals) at 37°C in 5% CO2. The pUASP vector was obtained from Drosophila Genomics Resource Center. A polyclonal rabbit antibody raised against full length PACT (produced in-house)29 was used at 1:8000 and was detected using 1:10000 horseradish peroxidase (HRP) conjugated polyclonal goat anti-rabbit secondary (Rockland Immunochemicals # 611-103-122).
Generation of pUASP plasmids
To obtain PACT cDNA, total RNA was extracted from HeLa cells using Trizol (Invitrogen) according to the manufacturer’s instructions, followed by DNAse treatment using the DNA-free kit (Ambion). Extracted, DNase-treated RNA was then reverse transcribed with the Superscript III system (Invitrogen) using random hexamers. The resulting cDNA was used to amplify the coding sequence of PACT by PCR using the primers 5´BamHI-PACT(UAS) (AAG GAT CCA AAC ATG TCC CAG AGC AGG CAC C) and 3´XbaI-PACT (GGC GGA TCC TTA CTT TCT TTC TGC TAT TAT CTT TAA ATA C). The resulting product was digested with BamHI (New England Biolabs Inc. #R0136) and XbaI (New England Biolabs Inc. #R0145) and ligated into pUASP to generate pUASP-PACT. The full-length Drosophila r2d2 cDNA was amplified by PCR from a plasmid obtained from Drosophila Genome Research Center (cDNA #LD06392) using 5´BamHI-R2D2(UAS) (AAG GAT CCA AAC ATG GAT AAC AAG TCA GCC GTA TC) and 3´XbaI-R2D2 (AAA TCT AGA TTA AAT CAA CAT GGT GCG AAA ATA GTC TAT TAT ATG G). The resulting product was digested with BamHI and XbaI and ligated into pUASP to generate pUASP-R2D2. Final cloned plasmids were sequence verified using the primers listed above by the Cleveland Clinic Lerner Research Institute Genomics Core.
Protein isolation and western blot
Protein was isolated by homogenizing individual adult flies in Triton X-100 lysis buffer (20mM Tris-HCl pH 7.5, 150mM NaCl, 1% Triton X-100, 1mM EDTA, 5mM 2-mercaptoethanol, 10% glycerol, supplemented with Complete protease inhibitor and PhoSTOP (Roche)). Protein was separated by sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membrane (Immobilon, Millpore) for western blot analysis. Western blots were visualized using enhanced chemiluminescence (ECL) detection reagents (GE Healthcare Lifesciences #RPN2106).
Drosophila strains and genetics
Flies were maintained and crossed at 25°C according to standard protocols30. Fly food was prepared according to the standard recipe from Bloomington Drosophila Stock Center with minor modifications (83.3ml/L molasses, 33.75g/L yeast, 97.58g/L cornmeal, 31g/L agar, 15.67ml/L Tegosept, all purchased from Genesee Scientific). Transgenic UAS-PACT and UAS-r2d2 flies were generated31 by Model Systems Genomics (Duke University, Durham, NC) using the plasmids pUASP-PACT and pUASP-R2D2 described above. The following fly strains were obtained from the Bloomington Drosophila Stock Center: w1118, r2d21/CyO (a null allele;27) and Df(2L)BSC142/CyO. Tubulin (tub)-GAL4 (insert on X) was a gift of Dr. A. Page-McCaw. Homozygous viable r2d21 flies were only obtained infrequently, suggesting that the stock acquired a second-site lethal mutation. Therefore, all experiments were performed by outcrossing r2d21 to the deficiency Df(2L)BSC142 (deletion of 28C3; 28D3, including the r2d2 gene at 28C4). Standard genetic methods were used to construct the following genotypes:1 tub-Gal4; r2d21/CyO;2 Df(2L)BSC142/CyO; UAS-PACT; and3 Df(2L)BSC142/CyO; UAS-r2d2.
Egg collection and hatch rate determination
Females of the genotype tub-GAL4; r2d21/Df(2L)BSC142; +/UAS-x (where x is PACT or r2d2) were outcrossed with w1118 males for fertility tests. Crosses to test fertility were established in vials of food for 24 hours prior to transferring flies to an egg collection chamber. Females were allowed to lay eggs on grape juice agar plates (Genesee Scientific, #47–102) for 1 hr prior to collecting the plate, and incubating at 25°C for 24 hours to allow eggs to hatch. The total number of eggs laid was determined by counting the number of larvae and the number of unhatched eggs. Hatch rate was calculated as the number of larvae divided by the number of total eggs laid. Student’s t-test of two independent experiments (defining P < 0.05 as statistically significant) was performed using Graphpad Prism version 6.02 software package (Graphpad Software).
Results
Generation of human PACT-expressing transgenic Drosophila melanogaster
We reasoned that the genetically tractable Drosophila model system could be used to study the in vivo functions of PACT. Because there is no known PKR ortholog in Drosophila, this also allows the role of PACT in small RNA processing to be investigated independent of PKR activation. Moreover, the well-characterized siRNA and miRNA processing pathways and availability of loqs and r2d2 mutant alleles make this an attractive system to study the role of PACT in the small RNA pathways. Various human proteins have been used to rescue Drosophila mutant phenotypes in order to identify homologous and orthologous proteins32–34, therefore to identify proteins orthologous to PACT, a human PACT transgene was constructed and introduced into Drosophila. This transgenic line was used to determine whether PACT can substitute for Drosophila dsRNA binding proteins that regulate small RNA processing (see below).
Human PACT-expressing D. melanogaster were created using the Gal4-UAS bipartite expression system35. This system allows the generation of flies containing transgenes of interest under the transcriptional control of the UAS element. These lines then can be crossed to a number of Gal4-driver flies to tailor transgene expression to the needs of a specific experiment. The full-length coding region of the Human PACT gene was cloned into the pUASP vector under the transcriptional control of the UASp element, a modified form of UAS that permits expression in both somatic and germline cells36. Several UAS-PACT transformants were obtained and their chromosomal insertions mapped. A stock that carried an insert on chromosome 3 was retained and crossed to the ubiquitous tub-GAL4 driver.
We next tested whether Human PACT was expressed at detectable levels in vivo. Flies expressing tub-Gal4 alone, containing the UAS-PACT transgene alone, and flies with tub-Gal4 driving UAS-PACT were analyzed by western blot for PACT expression in whole flies. Using an antibody that recognizes Human PACT29, we detected a protein band of the expected size (~34 kDa) in tub-GAL4; UAS-PACT flies but not in the individual parental lines (Figure 2). Importantly, because we obtained viable tub-GAL4; UAS-PACT adult flies, this experiment also demonstrates that ubiquitous transgenic expression of PACT does not impair Drosophila development.
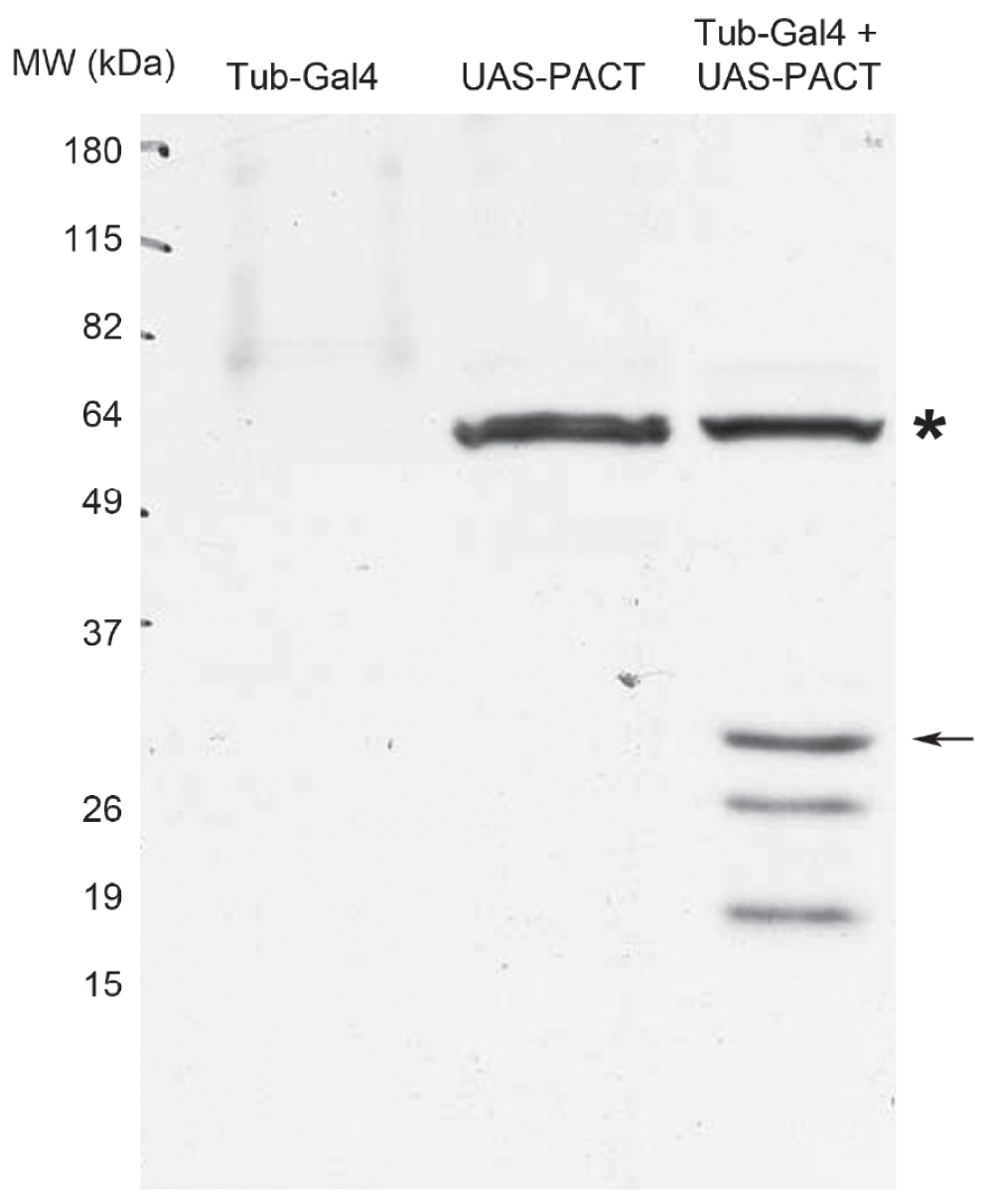
Figure 2. Expression of human PACT in transgenic Drosophila.
Western blot analysis of total protein isolated from whole flies of the indicated genotypes. Flies carrying both the tub-Gal4 and UAS-PACT elements show a protein band of expected size for PACT that is not present in either stock alone; a non-specific band is indicated (*). Arrow denotes predicted molecular weight of Human PACT (34 kDa). Additional bands of smaller size likely represent degradation products.
Expression of human PACT is insufficient to overcome the Drosophila R2D2 deficient phenotype
The focus of the work presented here is on PACT rescue of the r2d2 loss-of-function mutant phenotype. Investigation of the functional substitution between human PACT and Drosophila LOQS was investigated in a separate project in our lab and will be reported elsewhere.
Flies mutant for r2d2 have reduced fertility27. Therefore, we sought to determine the extent to which PACT can suppress r2d2 mutant fertility defects. We performed fertility tests using a standard egg-laying and hatching assay. Crosses were performed to obtain female progeny that carried the r2d21 allele over a deficiency that removes the r2d2 genetic locus (i.e. r2d2–/–), and simultaneously carried tub-GAL4 driving either UAS-PACT or UAS-r2d2 (Figure 3A; see Materials and methods for details). These female F1 progeny were then outcrossed to w1118 males to test for fertility; a cross of w1118 females to w1118 males was used as the wild-type control. Eggs resulting from these outcrosses were collected on grape juice agar plates, and allowed to hatch for 24 hours at 25°C. The total number of eggs produced and the number of hatched larvae were counted for each cross and hatch rate was calculated as the number of hatched larvae divided by the total number of eggs (Figure 3B & 3C). Flies heterozygous for r2d2 (either r2d21/+ or Df(2L)BSC142/+) produced eggs and hatch rates equivalent to the wild-type control. Notably, expression of either UAS-PACT or UAS-r2d2 in r2d2 heterozygous flies did not influence fertility or hatch rates. Consistent with a previous report27, flies homozygous mutant for r2d2 had a strong reduction in both the total number of eggs laid and the hatch rate compared to wild-type (Figure 3). We observed a rescue of both total number of eggs (p=0.0512) and hatch rate (p=0.0671) in r2d2 mutant flies by ubiquitous (tub-GAL4) transgenic expression of UAS-r2d2 that is approaching statistical significance and consistent with previous reports27. In contrast, ubiquitous expression of UAS-PACT did not suppress the homozygous r2d2 mutant fertility defects; the hatch rate and number of eggs laid was equivalent to the r2d2 mutants alone (Figure 3). Despite a documented interaction between PACT and the mammalian small RNA processing machinery and similar domain architecture between these proteins, these results demonstrate that Human PACT is not sufficient to rescue the R2D2 deficient phenotype.
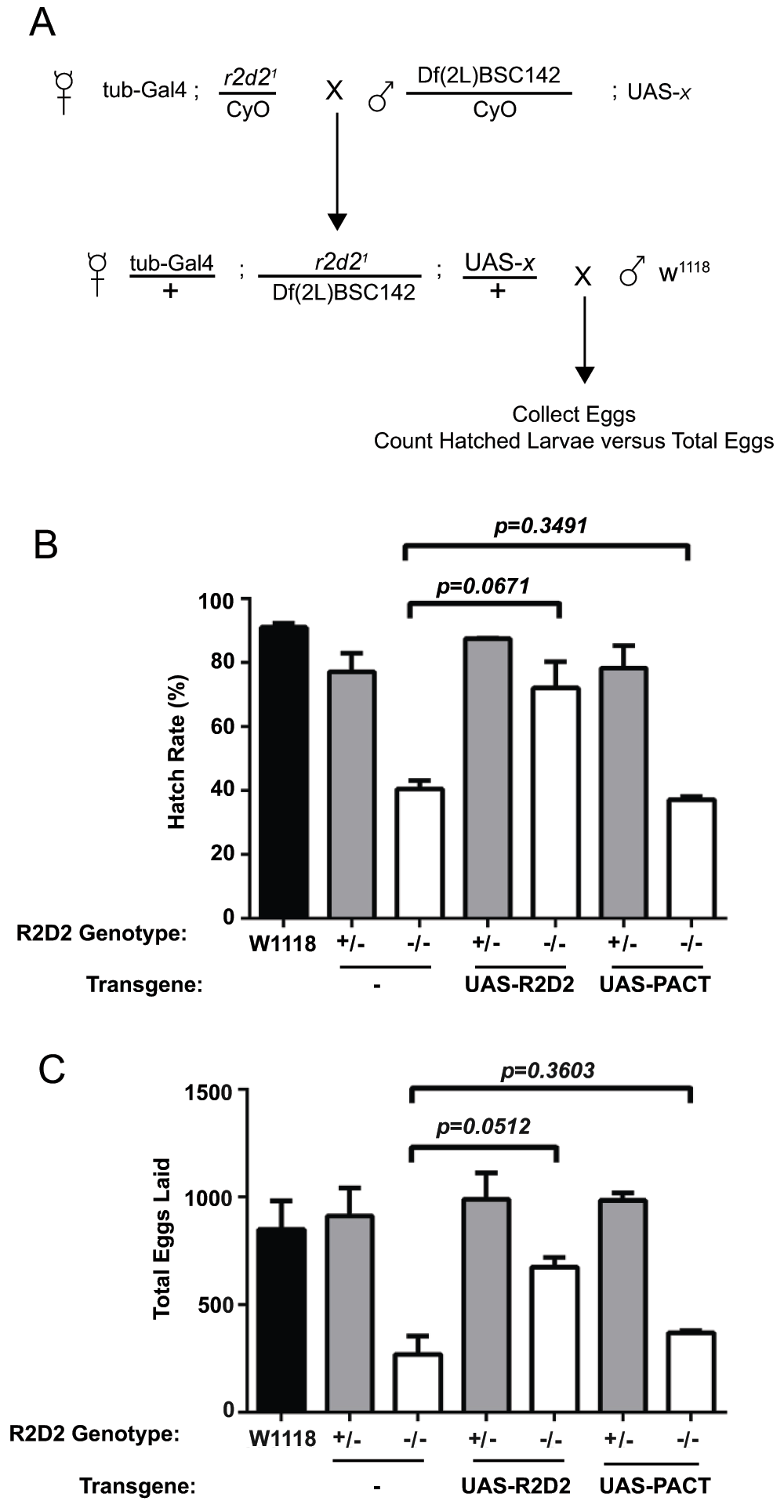
Figure 3. Rescue of the R2D2 deficient fertility defect by transgenic expression of R2D2 but not human PACT.
(A) Schematic representation of the experimental cross used to test fertility. UAS-x refers to the transgene shown in (B) and (C), for control crosses with no transgene the genotype Df(2L)BSC142/CyO; UAS-x was replaced with DF(2L)BSC142/CyO. (B) Calculated hatch rate of eggs produced by wild-type (w1118) flies, flies with one copy of r2d2 (+/-; either r2d21/CyO or Df(2L)BSC142/CyO) and flies lacking both copies of r2d2 (-/-, r2d21/Df(2L)BSC142) and either expressing no UAS-transgene or expressing UAS-PACT or UAS-r2d2; these flies were outcrossed to w1118 males for the fertility tests. (C) Total number of eggs laid from the crosses depicted in (B). Graphs show mean ± SEM of two independent experiments, p-values were calculated by Student’s t-test.
Discussion
Identification of PACT as a component of the small RNA processing pathway implicates this protein in multiple pathways of post-transcriptional gene regulation through general inhibition of translation by PKR and target-specific regulation by siRNA/miRNA. As such it is important to determine the precise nature of PACT’s involvement in this pathway in order to understand the overall function of PACT in post-transcriptional gene regulation, and the physiological effects of the protein. Here, we investigated the role of PACT in RNA silencing by attempting to identify orthologous proteins in D. melanogaster. Human PACT was introduced into Drosophila to determine whether it could substitute for R2D2, which has a well characterized function in RNA silencing. Expression of PACT was unable to rescue the phenotypes of flies lacking R2D2. These results suggest that the Dicer-associated dsRBPs PACT and R2D2 are not functionally orthologous.
While our study demonstrates that Human PACT cannot substitute for R2D2 function in flies, there are a number of important caveats to the results described here. Despite the lack of rescue, it is nonetheless possible that PACT plays the same (or a similar) role in mammalian cells as R2D2 plays in Drosophila cells. One possibility could be that PACT protein is sufficiently different from R2D2 that it cannot interact in vivo with Drosophila partner proteins that are necessary for R2D2 function. Alternatively, R2D2 may be required to bind specific RNA precursor molecules that PACT is unable to bind (or has a lower affinity for). Investigation of biochemical interactions between PACT and Drosophila RNA silencing proteins such as Dicer-1 and Dicer-2 and a biochemical comparison of the specific RNA binding functions of these proteins would begin to address these issues.
In addition to functional discrepancies between these proteins, there may be other potential explanations for these results that would not rule out orthologous function between PACT and R2D2. Although less likely, the lack of PACT rescue of the r2d2 mutant may be due to inherent difficulty in controlling the tissue-specificity and relative levels of transgene expression in vivo. In the case of R2D2, expression in the stalk and follicle cells of the ovary is required for egg development27. Expression of UAS-PACT by tub-GAL4 in whole flies was validated by western blot; however, this does not provide information about transgenic expression of PACT in the appropriate tissues. Thus, even though PACT expression was driven by a ubiquitous GAL4 driver we cannot rule out insufficiently high expression of PACT in the relevant cell types. Expression of UAS-R2D2 using the same tub-GAL4 driver however rescued the r2d2 loss-of-function fertility defect. Although this result provides some degree of confidence in the expression levels, the genomic context of the P-element insertion and stability of transgenically-produced RNA and protein can all contribute to expression variability in vivo. As such, it still remains a possibility that PACT was not expressed as highly as R2D2 using the same transgenic expression system, and thus PACT may not have rescued fertility due to insufficient expression rather than a lack of orthologous function. This issue could be addressed in future experiments by expressing epitope-tagged R2D2 or PACT in r2d2 mutant flies to correlate rescue of the r2d2 deficient phenotype with a direct comparison of protein expression level37. Additional experiments investigating tissue-specific expression levels of these proteins as well as using tissue-specific GAL4 drivers would begin to address these concerns.
While the complementation experiments described here indicate that PACT is unable to functionally substitute for R2D2, there are complications to the interpretation of these experiments. Further technical refinements will be necessary to definitively demonstrate or rule out functional orthology between PACT and R2D2. It also remains a distinct possibility that PACT is instead orthologous to LOQS, a topic that we are interested in investigating further in the future.
Author contributions
BKD contributed to the conception, design, execution, analysis and interpretation of the experiments detailed in this manuscript, as well as drafting and revising the manuscript. JAM contributed to the conception, design, analysis and interpretation of the experiments detailed in this manuscript, as well as revising the manuscript. GCS contributed to the conception, design, analysis and interpretation of the experiments detailed in this manuscript, as well as revising the manuscript.
Competing interests
No competing interests were disclosed.
Grant information
This work was supported by NIH grant CA062220, awarded to G. Sen.
Acknowledgments
We wish to thank George Aranjuez for technical assistance with Drosophila work.
Supplementary materials
Table S1. Hatch rate and egg count raw data replicate 1.
| Wild type | No transgene | UAS-R2D2 | UAS-PACT |
|---|
| W1118 | +/- | -/- | +/- | -/- | +/- | -/- |
|---|
| Larvae | 884 | 743 | 153 | 969 | 403 | 728 | 145 |
| Eggs laid | 983 | 1042 | 355 | 1112 | 631 | 1021 | 380 |
| % Hatched | 89.92879 | 71.30518 | 43.09859 | 87.14029 | 63.86688 | 71.30264 | 38.15789 |
Table S2. Hatch rate and egg count raw data replicate 2.
| Wild type | No transgene | UAS-R2D2 | UAS-PACT |
|---|
| W1118 | +/- | -/- | +/- | -/- | +/- | -/- |
|---|
| Larvae | 663 | 651 | 71 | 763 | 579 | 809 | 131 |
| Eggs laid | 718 | 785 | 187 | 870 | 721 | 948 | 361 |
| % Hatched | 92.33983 | 82.92994 | 37.96791 | 87.70115 | 80.30513 | 85.33755 | 36.28809 |
Faculty Opinions recommendedReferences
- 1.
Patel RC, Sen GC:
PACT, a protein activator of the interferon-induced protein kinase, PKR.
EMBO J.
1998; 17(15): 4379–4390. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 2.
Ito T, Yang M, May WS:
RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling.
J Biol Chem.
1999; 274(22): 15427–15432. PubMed Abstract
| Publisher Full Text
- 3.
Bennett RL, Blalock WL, Abtahi DM, et al.:
RAX, the PKR activator, sensitizes cells to inflammatory cytokines, serum withdrawal, chemotherapy, and viral infection.
Blood.
2006; 108(3): 821–829. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 4.
Lee ES, Yoon CH, Kim YS, et al.:
The double-strand RNA-dependent protein kinase PKR plays a significant role in a sustained ER stress-induced apoptosis.
FEBS Lett.
2007; 581(22): 4325–4332. PubMed Abstract
| Publisher Full Text
- 5.
Ruvolo PP, Gao F, Blalock WL, et al.:
Ceramide regulates protein synthesis by a novel mechanism involving the cellular PKR activator RAX.
J Biol Chem.
2001; 276(15): 11754–11758. PubMed Abstract
| Publisher Full Text
- 6.
Farrell PJ, Sen GC, Dubois MF, et al.:
Interferon action: two distinct pathways for inhibition of protein synthesis by double-stranded RNA.
Proc Natl Acad Sci U S A.
1978; 75(12): 5893–5897. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 7.
Dickerman BK, White CL, Chevalier C, et al.:
Missense mutation in the second RNA binding domain reveals a role for Prkra (PACT/RAX) during skull development.
PLoS One.
2011; 6(12): e28537. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 8.
Rowe TM, Rizzi M, Hirose K, et al.:
A role of the double-stranded RNA-binding protein PACT in mouse ear development and hearing.
Proc Natl Acad Sci U S A.
2006; 103(15): 5823–5828. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 9.
Peters GA, Seachrist DD, Keri RA, et al.:
The double-stranded RNA-binding protein, PACT, is required for postnatal anterior pituitary proliferation.
Proc Natl Acad Sci U S A.
2009; 106(26): 10696–10701. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 10.
Lee Y, Hur I, Park SY, et al.:
The role of PACT in the RNA silencing pathway.
EMBO J.
2006; 25(3): 522–532. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 11.
Kok KH, Ng MH, Ching YP, et al.:
Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA.
J Biol Chem.
2007; 282(24): 17649–17657. PubMed Abstract
| Publisher Full Text
- 12.
van Kouwenhove M, Kedde M, Agami R:
MicroRNA regulation by RNA-binding proteins and its implications for cancer.
Nat Rev Cancer.
2011; 11(9): 644–656. PubMed Abstract
| Publisher Full Text
- 13.
Czech B, Hannon GJ:
Small RNA sorting: matchmaking for Argonautes.
Nat Rev Genet.
2011; 12(1): 19–31. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 14.
MacRae IJ, Ma E, Zhou M, et al.:
In vitro reconstitution of the human RISC-loading complex.
Proc Natl Acad Sci U S A.
2008; 105(2): 512–517. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 15.
Gregory RI, Yan KP, Amuthan G, et al.:
The Microprocessor complex mediates the genesis of microRNAs.
Nature.
2004; 432(7014): 235–240. PubMed Abstract
| Publisher Full Text
- 16.
Tabara H, Yigit E, Siomi H, et al.:
The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans.
Cell.
2002; 109(7): 861–871. PubMed Abstract
| Publisher Full Text
- 17.
Saito K, Ishizuka A, Siomi H, et al.:
Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells.
PLoS Biol.
2005; 3(7): e235. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 18.
Liu Q, Rand TA, Kalidas S, et al.:
R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway.
Science.
2003; 301(5641): 1921–1925. PubMed Abstract
| Publisher Full Text
- 19.
Laraki G, Clerzius G, Daher A, et al.:
Interactions between the double-stranded RNA-binding proteins TRBP and PACT define the Medipal domain that mediates protein-protein interactions.
RNA Biol.
2008; 5(2): 92–103. PubMed Abstract
| Publisher Full Text
- 20.
Bernstein E, Caudy AA, Hammond SM, et al.:
Role for a bidentate ribonuclease in the initiation step of RNA interference.
Nature.
2001; 409(6818): 363–366. PubMed Abstract
| Publisher Full Text
- 21.
Förstemann K, Tomari Y, Du T, et al.:
Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein.
PLoS Biol.
2005; 3(7): e236. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 22.
Zhou R, Czech B, Brennecke J, et al.:
Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform.
RNA.
2009; 15(10): 1886–1895. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 23.
Tomari Y, Du T, Zamore PD:
Sorting of Drosophila small silencing RNAs.
Cell.
2007; 130(2): 299–308. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 24.
Marques JT, Kim K, Wu PH, et al.:
Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila.
Nat Struct Mol Biol.
2010; 17(1): 24–30. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 25.
Liu X, Jiang F, Kalidas S, et al.:
Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes.
RNA.
2006; 12(8): 1514–1520. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 26.
Park JK, Liu X, Strauss TJ, et al.:
The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells.
Curr Biol.
2007; 17(6): 533–538. PubMed Abstract
| Publisher Full Text
- 27.
Kalidas S, Sanders C, Ye X, et al.:
Drosophila R2D2 mediates follicle formation in somatic tissues through interactions with Dicer-1.
Mech Dev.
2008; 125(5–6): 475–485. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 28.
Horne-Badovinac S, Bilder D:
Mass Transit: epithelial morphogenesis in the Drosophila egg chamber.
Dev Dyn.
2005; 232(3): 559–574. PubMed Abstract
| Publisher Full Text
- 29.
Marques JT, White CL, Peters GA, et al.:
The role of PACT in mediating gene induction, PKR activation and apoptosis in response to diverse stimuli.
J Interferon Cytokin Res.
2008; 28(8): 469–76. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 30.
Greenspan RJ:
Fly pushing: the theory and practice of Drosophila genetics. 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. 2004. Reference Source
- 31.
Bachmann A, Knust E:
The use of P-element transposons to generate transgenic flies.
Methods Mol Biol.
2008; 420: 61–77. PubMed Abstract
| Publisher Full Text
- 32.
Ahern-Djamali SM, Comer AR, Bachmann C, et al.:
Mutations in Drosophila enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains.
Mol Biol Cell.
1998; 9(8): 2157–2171. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 33.
Grifoni D, Garoia F, Schimanski CC, et al.:
The human protein Hugl-1 substitutes for Drosophila lethal giant larvae tumour suppressor function in vivo.
Oncogene.
2004; 23(53): 8688–8694. PubMed Abstract
| Publisher Full Text
- 34.
Ryan EL, DuBoff B, Feany MB, et al.:
Mediators of a long-term movement abnormality in a Drosophila melanogaster model of classic galactosemia.
Dis Model Mech.
2012; 5(6): 796–803. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 35.
Brand AH, Perrimon N:
Targeted gene expression as a means of altering cell fates and generating dominant phenotypes.
Development.
1993; 118(2): 401–415. PubMed Abstract
- 36.
Rørth P:
Gal4 in the Drosophila female germline.
Mech Dev.
1998; 78(1–2): 113–118. PubMed Abstract
| Publisher Full Text
- 37.
Corbett A:
Referee Report For: The Human dsRNA binding protein PACT is unable to functionally substitute for the Drosophila dsRNA binding protein R2D2 [v1; ref status: indexed].
F1000Research.
2014; 2: 220. Publisher Full Text



Comments on this article Comments (0)