Introduction
Throughout the history of medicine there has been an awareness of animal to human transmission of disease, and the etiological pathogens have been collectively described as zoonoses1. Water fowl and wild birds have been identified as reservoirs for the virus Influenza A2,3; a highly mutable and infectious pathogen that infects avian and mammalian species4. Ducks are observed in a multitude of fresh water sources including ponds, water fountains and pools where they can defecate; bacteria have been shown to be distributed through aerosols from ornamental fountains5,6 and reclaimed water dispensed through an irrigation system7. Humans may also have direct contact with ducks and their excrement through the recreational sport of duck hunting8. Ducks can also shed pathogens near chicken farms or other animals—such as pigs—that have access to outside areas. An avian influenza A virus (H7N7) epidemic in the Netherlands in 2003 thought to be initiated from a migratory water fowl resulted in the culling of 30 million poultry in an area of the country where free-range poultry farming was common9. Due to the migratory nature and unrestrained behavior of the wild duck, Aythya Americana, our study set out to investigate the bacterial microbiome of a wild duck and to identify its bacterial flora relative to the same bacterial species that have been reported to cause disease in farm animals and humans.
Methods
Amplicon pyrosequencing (bTEFAP) was originally described by Dowd et al.10 and has been used in describing a wide range of environmental and health related microbiomes including the intestinal populations of a variety of animals and their environments including cattle11–15. A fecal sample obtained from a wild duck, Aythya americana, that was killed during duck hunting season (December 2012) by a licensed hunter, was aseptically swabbed onto a Whatman FTA card (GE Healthcare Life Sciences) using a sterile swab and gloves being careful to avoid environmental contamination. The flap of the FTA card was placed over the FTA paper and placed into a sterile pouch, and the FTA card was stored at room temperature prior to DNA amplification. 2 mm punches were washed with FTA reagent and TE (10 mM Tris-HCL, 1 mM EDTA, pH 8.0) according to the manufacturer’s protocol, and the dried punches were used as template DNA for thermal cycling. DNA was also isolated from pond water as a negative comparison and sampled from a source of water visited by numerous avian species but not at the source of the fecal sampling but within the migratory range of Aythya americana. The pond water DNA was isolated using water RNA/DNA purification kit (0.45 µm) [Norgen Biotek Corp, Thorold, ON, Canada]. For thermal cycling and DNA amplification we used the 16S universal Eubacterial primers 27f 5´-AGAGTTTGATCCTGGCTCAG-3´ and 1492r primer 5´-ACGGCTACCTTGTTACGACTT-3´ (Integrated DNA Technologies). A single-step 30 cycle PCR using EconoTaq PLUS 2X Master Mix (Lucigen, Middleton, WI) were used under the following conditions: 94°C for 2 minutes, followed by 30 cycles of 95°C for 120 seconds; 42°C for 30 seconds and 72°C for 4 minutes; after which a final elongation step at 72°C for 20 minutes was performed. Following PCR, DNA products were resolved in a 1% agarose, 1X TAE gel stained with ethidium bromide and 1.5 Kb products were excised from the gel purified using a cyclo-prep spin column (Amresco, Solon, OH). All the DNA products were purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, MA, USA). Samples were sequenced using Roche 454 FLX titanium instruments and reagents following manufacturer’s guidelines. The Q25 sequence data derived from the sequencing process was processed using a proprietary analysis pipeline (www.mrdnalab.com, MR DNA, Shallowater, TX). Sequences were depleted of barcodes and primers. Next, short sequences < 200bp, sequences with ambiguous base calls, and sequences with homopolymer runs exceeding 6bp were removed. Sequences were then denoised and chimeras removed. Operational taxonomic units (OTUs) were defined after removal of singleton sequences, clustering at 3% divergence--97% similarity10,15. OTUs were then taxonomically classified using BLASTn against a curated GreenGenes database16,17 and compiled into each taxonomic level into both “counts” and “percentage” files.
Results
Due to the aquatic nature of the animal, we initially expected that the biodiversity of bacterial species in the duck feces would reflect numerous bacterial species present in the pond water, and since we observed multiple species of aquatic birds in the pond we expected to find eubacteria in common. Figure 1 is a modified heat map showing differences and similarities among the classes of eubacteria sequenced and identified. The figure demonstrates clear differences at the taxonomical level of Class with few common classes of bacteria namely Actinobacteria, Clostridia and Gammaproteobactera.
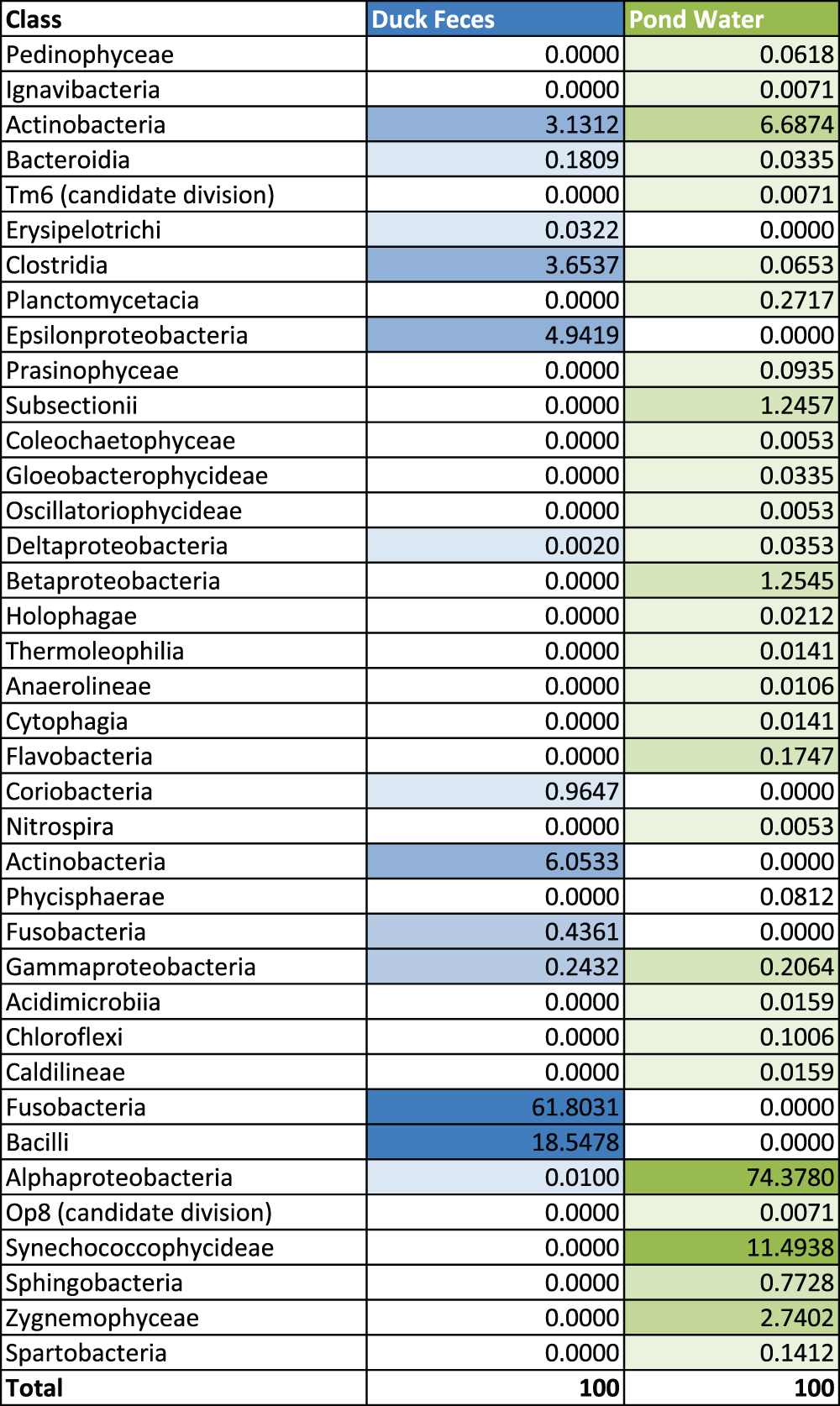
Figure 1. Comparison of Classes of Eubacteria present in the Duck to the Classes of Eubacteria present in pond water using a modified heat map.
Darker colors represent a higher representation of the bacterial class.
However, similarities at the level of Genus and species included only Agrobacterium tumefaciencs and a species of Porphyromonas and a species of Ruminococcaceae (Figure 2). This analysis indicated distinct differences between the eubacteria present in the duck fecal sample and the pond water sample, and it also indicated that our sampling of the duck feces was devoid of any obvious pond water eubacterial constituents.
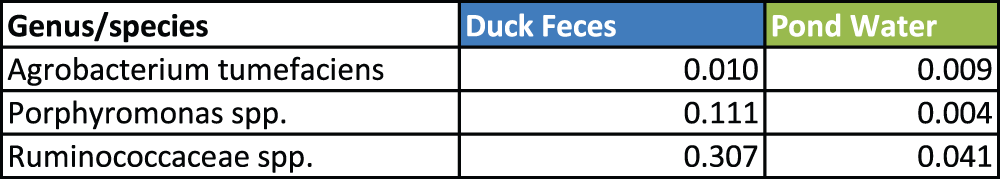
Figure 2. Bacterial species present in both duck feces and pond water.
The taxonomical classification of OTU at the level of genus and species was compiled in relation to percentages of the Eubacterial microbiome (Table 1). In Table 2, we referenced reported cases of diseases related to the bacteria sequenced from the duck’s fecal sample reflecting the eubacterial microbiome’s potential to cause disease in humans and other mammals. The largest representative bacterial species—relative to percentage—was Fusobacterium mortiferum at 31.6%. Fusobacterium mortiferum reports related to human disease are sparse, but Fusobacterium have been associated with rare but serious cases of bacteremia18,19, and a 6 year study of “other gram-negative anaerobic bacilli” (OGNAB) isolated from anaerobic infections at the Wadsworth Clinical Anaerobic Clinical Anaerobic Bacteriology Research Laboratory in Los Angeles, CA reported that most strains of Fusobacteria—outside of Fusobacterium nucleatum—were resistant to erythromycin20. The pathogen, Fusobacterium nucleatum, on the other hand, is well-known for its association with disease and its ability to adhere to Gram-positive and Gram-negative bacteria in dental biofilms such as plaque21.
Table 1. Taxonomical classification of operational taxonomic units into the Genus/species level with representative percentages of the Eubacterial Microbiome.
| OTUs genus/species | % of Eubacterial microbiome |
|---|
| Fusobacterium mortiferum | 31.609 |
| Streptobacillus moniliformis | 30.100 |
| Lactobacillus intermedius | 11.021 |
| Actinomyces suimastitidis | 4.474 |
| Campylobacter canadensis | 3.694 |
| Enterococcus cecorum | 3.585 |
| Lactobacillus aviarius | 2.792 |
| Actinomyces spp. | 1.966 |
| Pseudobutyrivibrio spp. | 1.811 |
| Helicobacter brantae | 1.248 |
| Coriobacteriaceae spp. | 0.928 |
| Actinomyces nasicola | 0.784 |
| Actinomyces odontolyticus | 0.699 |
| Lactobacillus aviarius | 0.627 |
| Roseburia spp. | 0.380 |
| Leptotrichia spp. | 0.364 |
| Ruminococcaceae spp. | 0.307 |
| Actinomyces turicensis | 0.295 |
| Bacillus spp. | 0.265 |
| Plesiomonas shigelloides | 0.239 |
| Fastidiosipila spp. | 0.213 |
| Actinomyces canis | 0.209 |
| Arcanobacterium pyogenes | 0.183 |
| Blautia spp. | 0.169 |
| Ruminococcus spp. | 0.157 |
| Veillonella ratti | 0.155 |
| Actinomyces europaeus | 0.139 |
| Atopobium vaginae | 0.121 |
| Lactobacillus spp. | 0.117 |
| Porphyromonas spp. | 0.111 |
| Parvimonas micra | 0.078 |
| Tessaracoccus spp. | 0.068 |
| Fusobacterium periodonticum | 0.058 |
| Atopobium rimae | 0.054 |
| Oscillibacter spp. | 0.054 |
| Helcococcus kunzii | 0.054 |
| Arthrobacter bergerei | 0.048 |
| Streptococcus macedonicus | 0.044 |
| Clostridium spp. | 0.044 |
| Peptostreptococcaceae spp. | 0.042 |
| Enterococcus spp. | 0.040 |
| Cetobacterium ceti | 0.038 |
| Veillonella magna | 0.036 |
| Cetobacterium spp. | 0.034 |
| Peptoniphilus asaccharolyticus | 0.034 |
| Flavonifractor spp. | 0.034 |
| Fusobacterium nucleatum | 0.030 |
| Actinomyces neuii | 0.026 |
| Bacteroides plebeius | 0.024 |
| Veillonella dispar | 0.024 |
| Streptococcus spp. | 0.020 |
| Dorea spp. | 0.018 |
| Allobaculum spp. | 0.016 |
| Porphyromonas gingivalis | 0.016 |
| Eubacterium sulci | 0.016 |
| Actinomyces lingnae | 0.016 |
| Bacteroides spp. | 0.016 |
| Collinsella spp. | 0.016 |
| Actinoplanes roseosporangius | 0.014 |
| Erysipelotrichaceae spp. | 0.014 |
| Lysinibacillus spp. | 0.014 |
| Corynebacterium freneyi | 0.014 |
| Myceligenerans xiligouense | 0.012 |
| Actinomyces vaccimaxillae | 0.012 |
| Streptococcus suis | 0.012 |
| Anaerotruncus spp. | 0.012 |
| Sporosarcina spp. | 0.010 |
| Isoptericola variabilis | 0.010 |
| Olsenella spp. | 0.010 |
| Atopobium spp. | 0.010 |
| Agrobacterium tumefaciens | 0.010 |
| Microbispora rosea | 0.008 |
| Actinocorallia glomerata | 0.008 |
| Coprococcus spp. | 0.008 |
| Mobiluncus curtisii | 0.008 |
| Bacteroides coprocola | 0.008 |
| Prevotellaceae spp. | 0.006 |
| Sneathia spp. | 0.006 |
| Veillonella spp. | 0.006 |
| Gardnerella spp. | 0.006 |
| Varibaculum cambriense | 0.006 |
| Acinetobacter spp. | 0.004 |
| Actinomyces hongkongensis | 0.004 |
| Turicibacter spp. | 0.002 |
| Desulfovibrio spp. | 0.002 |
| Total | 100 |
Table 2. Diseases related to the eubacteria identified in the wild duck fecal microbiome.
| Genus/Species | % of Biome | Disease | Reference |
|---|
| Fusobacterium mortiferum | 31.61 | Septicemia | 18–20 |
| Streptobacillus moniliformis | 30.10 | Rat bite fever/Haverhill, osteomyelitis, epidural abscess, fever and polyarthralgia, bacteremia, drinking water related disease | 22–27 |
| Lactobacillus intermedius | 11.02 | Renal transplant infection | 28 |
| Actinomyces suimastitidis | 4.47 | Pig mastitis | 29 |
| Campylobacter canadensis | 3.69 | Drinking water related disease | 27 |
| Enterococcus cecorum | 3.59 | Arthritis and osteomyelitis in chicks, enteroccocal spondylitis (ES) chicks, Aortic valve endocarditis in humans, empyema thoracis, septicemia, recurrent bacteremic peritonitis | 30–35 |
| Actinomyces odontolyticus | 0.70 | Bacteremia in immunosuppressed patients, Malar mass | 36,37 |
| Leptotrichia spp. | 0.36 | Bacteremia | 38 |
| Actinomyces turicensis | 0.30 | Genital infections, urinary infections, skin infections, post-operative wound infection, abscess, appendicitis, ear and nose and throat infection, and bacteremia | 39 |
| Plesiomonas shigelloides | 0.24 | Travelers' diarrhea, dysentery, gastroenteritis | 45–48 |
| Arcanobacterium pyogenes | 0.18 | Human wound infections | 49 |
| Actinomyces europaeus | 0.14 | Human abscesses | 40 |
| Atopobium vaginae | 0.12 | Bacterial vaginosis | 50 |
| Parvimonas micra | 0.08 | Odontogenic infection | 52 |
| Atopobium rimae | 0.05 | Human Bacteremia | 53 |
| Helcococcus kunzii | 0.05 | Urocystitis in a sow | 59 |
| Fusobacterium nucleatum | 0.03 | Bacteremia | 54 |
| Actinomyces neuii | 0.03 | Endophthalmitis, periprosthetic infection | 41,42 |
| Veillonella dispar | 0.02 | Septic arthritis | 57 |
| Porphyromonas gingivalis | 0.02 | Periodontitis | 32 |
| Corynebacterium freneyi | 0.01 | Bacteremia | 55 |
| Actinomyces vaccimaxillae | 0.01 | Cow jaw lesion | 43 |
| Streptococcus suis | 0.01 | Meningitis, septicemia, endocarditis, arthritis, and septic shock in both pigs and human beings, | 56 |
| Varibaculum cambriense | 0.01 | Intrauterine devices and vagina | 51 |
| Actinomyces hongkongensis | <0.01 | Bacteremia | 44 |
Streptobacillus moniliformis was also identified as a major constituent of the duck fecal eubacterial microbiome at 30.1%. Several well-studied and documented cases of disease are attributed to S. moniliformis including rat bite fever or Haverhill disease22, osteomyelitis23, epidural abscesses24, fever and polyarthralgia25, bacteremia26 and contaminated drinking water related disease27.
Other organisms and their respective illnesses included Lactobacillus intermedius (11.02%) in a renal transplant infection28, Actinomyces suimastitidis (4.47%) in pig mastitis29 and Campylobacter canadensis (3.69%) in drinking water related disease27. Enterococcus cecorum was another identified pathogen at 3.59% of the sequenced Eubacterial microbiome, and E. cecorum has been reported to cause disease in chicks30,31 and humans including aortic valve endocardititis32, empyema thoracis33, septicemia in a malnourished adult34 and recurrent bacteremic peritonitis in a patient with liver cirrhosis35. Actinomyces odontolyticus (0.70%) has recently been reported to cause bacteremia in immunosuppressed patients36, and members of the genus Actinomyces have been known to cause actinomycosis for some time. A. odonolyticus was reported by Michell, Hintz and Haselby in 1997 to be the cause of a malar mass in soft tissue in a human37. A species of the genus Leptotrichia (0.36%) was also identified, a genus that has been associated with bacteremia in multiple myeloma patients receiving chemotherapy38. Another Actinomyces present in the wild duck eubacterial microbiome was Actinomyces turicensis at 0.3%, a bacterium associated with a spectrum of diseases including genital infections, urinary tract infections, skin infections, post-operative wound infection, abscesses, appendicitis, ear and nose and throat infection and bacteremia39. In addition, Actinomyces europaeus (0.14%) was reported in human abscesses40, Actinomyces neuii (0.03%) was reported to cause endophthalmitis41 and periprosthetic infection42, Actinomyces vaccimaxillae (0.01%) was isolated from a cow jaw lesion43 and Actinomyces hongkongensis (0.004%) was reported to cause high-mortality bacteremia in humans44.
0.24% of the eubacterial population was composed of Plesiomonas shigelloides a well-documented pathogen associated with Travelers’ diarrhea, dysentery and gastroenteritis45–48. Arcanobacterium pyogenes was also present (0.18%), a pathogen reported to cause soft tissue infections in humans49. Atopobium vaginae (0.12%) was reported to cause bacterial vaginosis in a human50 and Varibaculum cambriense (0.01%) was reported to cause complications with intrauterine devices and vaginal infections in Hong Kong51, Parvimonas micra (0.08%) was associated with odontogenic infection52 and human bacteremia was reported with Atopobium rima53, Fusobacterium nucleatum54, Corynebacterium freneyi55 and Streptococcus suis56. Finally, Veillonella dispar (0.02%) was reported in a case of septic arthritis57 and Porphyromonas gingivalis (0.02%) is a well-studied pathogen reported decades earlier and associated with periodontitis58.
Discussion
Numerous pathogenic eubacterial species have been identified in the fecal sample obtained from the wild duck, Aythya Americana, using amplicon pyrosequencing, a widely accepted method for analyzing the bacterial composition of microbial ecosystems. We were surprised to find that most of the species of eubacteria sequenced the duck feces were not present in a pond water sample from a water source that was known to be visited by numerous water fowl. Perhaps, the analyses of small samples from a pond or lake are not adequate when investigating the presence of avian contamination.
The summary in Table 2 indicates that many of the bacteria that are listed are clinically important causing severe diseases such as bacteremia and septicemia. The potential to cause disease can be appreciated when one considers that wild-duck feces can contaminate food, drinking water and open wounds. In addition, bird feces can easily contaminate ornamental fountains--where aerosols are produced—and the aerosols can carry the bacteria in a similar way to what has been reported for Legionella pneumophila47. It is possible that many of the bacterial entities when disseminated to humans and other animals could also cause subclinical respiratory illnesses that are not reported due to patient resolution.
It is only prudent to recommend that immunocompromised humans and animals should limit their exposure to environments where ducks may have polluted the water source—this includes outdoor pools and fountains. That realization also supports the practice of adequately chlorinating or sanitizing artificial pools and fountains to prevent opportunistic infections through aerosols or breaks in the skin. Duck hunters should also be aware of the risk of bacterial contamination in addition to the risk posed by the influenza virus. Additionally, reclaimed water poses a threat to the elderly and other immunocompromised humans who might be exposed to aerosols that are produced when the reclaimed water is used as a source of irrigation such as in golf courses and gardens, a common practice that might warrant further inquiry.
When determining the cause of disease it is difficult—if not impossible—to identify the source of infection, and whether it has indeed originated from an animal that is migratory or aquatic in nature. Many of the bacterial species that were cited to cause infections among humans were also found in the excrement of a migratory and aquatic bird that defecates in water supplies and around other animals. However, since our analysis was limited to the careful analysis of a single, wild duck’s eubacterial microbiome, the disease potential was relative to that animal only and cannot be extrapolated to all ducks of the same species. Thus, the disease potential is relative to this studied microbiome and further statistical studies will be needed to determine the global risks associated with duck excrement among different species.
Author contributions
TS carried out the majority of the molecular biology techniques in the laboratory, AG was instrumental in obtaining the wild ducks specimens. SD provided expertise in pyrosequencing and bioinformatics. JC conceived the study and wrote the first draft of the manuscript. All authors were involved in the revision of the draft and have agreed to the final content. The study was an active learning exercise that helped bridge the understanding of Medical Microbiology with field research, molecular biology and bioinformatics for graduate students seeking their Masters (MS) degree in Biomedical Sciences under the guidance of Dr. Coffman.
Competing interests
No competing interests were disclosed.
Grant information
The work was funded by the Barry University Faculty Incentive Grant 10-110106 awarded to Dr. Jonathan Coffman.
Acknowledgements
We would like to acknowledge Barry University for providing the infrastructure to conduct the experiments.
Faculty Opinions recommendedReferences
- 1.
Spickler AR, Roth JA, Galyon J, et al.:
Emerging and Exotic Diseases of Animals Textbook. 4th ed. Center for Food Security and Public Heatlh. 2010; 383. Reference Source
- 2.
Alexander DJ:
A review of avian influenza in different bird species.
Vet Microbiol.
2000; 74(1–2): 3–13. PubMed Abstract
| Publisher Full Text
- 3.
Bahl J, Krauss S, Kuhnert D, et al.:
Influenza a virus migration and persistence in north american wild birds.
PLoS Pathog.
2013; 9(8): e1003570. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 4.
Kocer ZA, Krauss S, Stallknecht DE, et al.:
The potential of avian H1N1 influenza A viruses to replicate and cause disease in mammalian models.
PLoS One.
2012; 7(7): e41609. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 5.
O’Loughlin RE, Kightlinger L, Werpy MC, et al.:
Restaurant outbreak of Legionnaires’ disease associated with a decorative fountain: an environmental and case-control study.
BMC Infect Dis.
2007; 7(1): 93–99. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 6.
Escartin EF, Lozano JS, Garcia OR, et al.:
Potential Salmonella transmission from ornamental fountains.
J Environ Health.
2002; 65(4): 9–12. PubMed Abstract
- 7.
Herzog A, Bhaduri P, Stedtfeld RD, et al.:
Detection and occurrence of indicator organisms and pathogens.
Water Environ Res.
2010; 82(10): 883–907. Publisher Full Text
- 8.
Dishman H, Stallknecht D, Cole D:
Duck hunters’ perceptions of risk for avian influenza, Georgia, USA.
Emerg Infect Dis.
2010; 16(8): 1279–1281. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 9.
Stegeman A, Bouma A, Elbers AR, et al.:
Avian influenza A virus (H7N7) epidemic in The Netherlands in 2003: course of the epidemic and effectiveness of control measures.
J Infect Dis.
2004; 190(12): 2088–2095. PubMed Abstract
| Publisher Full Text
- 10.
Dowd SE, Callaway TR, Wolcott RD, et al.:
Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP).
BMC Microbiol.
2008; 8: 125. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 11.
Caporaso JG, Lauber CL, Walters WA, et al.:
Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample.
Proc Natl Acad Sci U S A.
2011; 108(Suppl 1): 4516–4522. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 12.
Capone KA, Dowd SE, Stamatas GN, et al.:
Diversity of the human skin microbiome early in life.
J Invest Dermatol.
2011; 131(10): 2026–2032. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 13.
Dowd SE, Sun Y, Wolcott RD, et al.:
Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs.
Foodborne Pathog Dis.
2008; 5(4): 459–472. PubMed Abstract
| Publisher Full Text
- 14.
Eren AM, Zozaya M, Taylor CM, et al.:
Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation.
PLoS One.
2011; 6(10): e26732. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 15.
Dowd SE, Hanson JD, Rees E, et al.:
Survey of fungi and yeast in polymicrobial infections in chronic wounds.
J Wound Care.
2011; 20(1): 40–47. PubMed Abstract
- 16.
DeSantis TZ, Hugenholtz P, Larsen N, et al.:
Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB.
Appl Enviro Micro.
2006; 72(7): 5069–5072. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 17.
Edgar RC:
Search and clustering orders of magnitude faster than BLAST.
Bioinformatics.
2010; 26(19): 2460–2461. PubMed Abstract
| Publisher Full Text
- 18.
Afra K, Laupland K, Leal J, et al.:
Incidence, risk factors, and outcomes of Fusobacterium species bacteremia.
BMC Inf Dis.
2013; 13(1): 264. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 19.
Prout J, Glymph R:
Anaerobic septicemia secondary to Fusobacterium mortiferum.
J Natl Med Assoc.
1986; 78(4): 334–337. PubMed Abstract
| Free Full Text
- 20.
George WL, Kirby BD, Sutter VL, et al.:
Gram-negative anaerobic bacilli: their role in infection and patterns of susceptibility to antimicrobial agents. II. Little-known Fusobacterium species and miscellaneous genera.
Rev Infect Dis.
1981; 3(3): 599–626. PubMed Abstract
- 21.
Van der Velden U, Van Winkelhoff AJ, Abbas F, et al.:
The habitat of periodontopathic micro-organisms.
J Clin Periodontol.
1986; 13(3): 243–248. PubMed Abstract
| Publisher Full Text
- 22.
Himsworth CG, Parsons KL, Jardine C, et al.:
Rats, cities, people, and pathogens: a systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers.
Vector Borne Zoonotic Dis.
2013; 13(6): 349–359. PubMed Abstract
| Publisher Full Text
- 23.
Flannery DD, Akinboyo I, Ty JM, et al.:
Septic arthritis and concern for osteomyelitis in a child with rat bite fever.
J Clin Microbiol.
2013; 51(6): 1987–1989. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 24.
Addidle M, Pynn J, Grimwade K, et al.:
Epidural abscess caused by Streptobacillus moniliformis.
J Clin Microbiol.
2012; 50(9): 3122–3124. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 25.
Orlev A, Miskin I, Temper V, et al.:
A 70-year-old man with fever and polyarthralgia. Streptobacillus moniliformis.
Clin Infect Dis.
2011; 53(10): 1037–1038. PubMed Abstract
| Publisher Full Text
- 26.
Joshi RM, Al Sweih N, Bin Nakhi HA, et al.:
Streptobacillus moniliformis bacteremia in a child: case report.
Med Princ Pract.
2010; 19(5): 409–411. PubMed Abstract
| Publisher Full Text
- 27.
Nichols G, Lane C, Asgari N, et al.:
Rainfall and outbreaks of drinking water related disease and in England and Wales.
J Water Health.
2009; 7(1): 1–8. PubMed Abstract
| Publisher Full Text
- 28.
Domann E, Hong G, Imirzalioglu C, et al.:
Culture-independent identification of pathogenic bacteria and polymicrobial infections in the genitourinary tract of renal transplant recipients.
J Clin Microbiol.
2003; 41(12): 5500–5510. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 29.
Hoyles L, Falsen E, Holmstrom G, et al.:
Actinomyces suimastitidis sp. nov., isolated from pig mastitis.
Int J Syst Evol Microbiol.
2001; 51(4): 1323–1326. PubMed Abstract
| Publisher Full Text
- 30.
Stalker MJ, Brash ML, Weisz A, et al.:
Arthritis and osteomyelitis associated with Enterococcus cecorum infection in broiler and broiler breeder chickens in Ontario, Canada.
J VET Diagn Invest.
2010; 22(4): 643–645. PubMed Abstract
| Publisher Full Text
- 31.
Borst LB, Suyemoto MM, Robbins KM, et al.:
Molecular epidemiology of Enterococcus cecorum isolates recovered from enterococcal spondylitis outbreaks in the southeastern United States.
Avian Pathol.
2012; 41(5): 479–485. PubMed Abstract
| Publisher Full Text
- 32.
Ahmed FZ, Baig MW, Gascoyne-Binzi D, et al.:
Enterococcus cecorum aortic valve endocarditis.
Diagn Microbiol Infect Dis.
2011; 70(4): 525–527. PubMed Abstract
| Publisher Full Text
- 33.
Woo PC, Tam DM, Lau SK, et al.:
Enterococcus cecorum empyema thoracis successfully treated with cefotaxime.
J Clin Micrbiol.
2004; 422): 919–922. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 34.
Greub G, Devriese LA, Pot B, et al.:
Enterococcus cecorum septicemia in a malnourished adult patient.
Eur J Clin Microbiol Infect Dis.
1997; 16(8): 594–598. PubMed Abstract
| Publisher Full Text
- 35.
Hsueh PR, Teng LJ, Chen YC, et al.:
Recurrent bacteremic peritonitis caused by Enterococcus cecorum in a patient with liver cirrhosis.
J Clin Microbiol.
2000; 38(6): 2450–2452. PubMed Abstract
| Free Full Text
- 36.
Cone LA, Leung MM, Hirschberg J:
Actinomyces Odontolyticus bacteremia.
Emerg Infect Dis.[serial online].
2003; 9(12): 1629–32. PubMed Abstract
| Publisher Full Text
- 37.
Mitchell PD, Hintz CS, Haselby RC:
Malar mass due to Actinomyces odontolyticus.
J Clin Microbiol.
1977; 5(6): 658–60. PubMed Abstract
| Free Full Text
- 38.
Couturier MR, Slechta ES, Goulston C, et al.:
Leptotrichia bacteremia in patients receiving high-dose chemotherapy.
J Clinical Micro.
2012; 50(4): 1228–1232. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 39.
Sabbe LJ, Van De Merwe D, Schouls L, et al.:
Clinical spectrum of infections due to the newly described Actinomyces species A. turicensis, A. radingae, and A. europaeus.
J Clin Micro.
1999; 37(1): 8–13. PubMed Abstract
| Free Full Text
- 40.
Funke G, Alvarez N, Pascual C, et al.:
Actinomyces europaeus sp. nov., isolated from human clinical specimens.
Int J Syst Bacter.
1997; 47(3): 687–692. PubMed Abstract
| Publisher Full Text
- 41.
Graffi S, Peretz A, Naftali M:
Endogenous endophthalmitis with an unusual infective agent: Actinomyces neuii.
Eur J Ophthalmol.
2012; 22(5): 834–835. PubMed Abstract
| Publisher Full Text
- 42.
Rieber H, Schwarz R, Kramer O, et al.:
Actinomyces neuii subsp. neuii associated with periprosthetic infection in total hip arthroplasty as causative agent.
J Clin Microbiol.
2009; 47(12): 4183–4184. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 43.
Hall V, Collin MD, Hutson R, et al.:
Actinomyces vaccimaxillae sp. nov., from the jaw of a cow.
Int J Syst Evol Microbiol.
2003; 53(Pt 2): 603–606. PubMed Abstract
| Publisher Full Text
- 44.
Lau SK, Fan RY, Lo HW, et al.:
High mortality associated with Catabacter hongkongensis bacteremia.
J Clin Microbiol.
2012; 50(7): 2239–2243. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 45.
Shah N, DuPont HL, Ramsey DJ:
Global etiology of travelers’ diarrhea: systematic review from 1973 to the present.
Am J Trop Med Hyg.
2009; 80(4): 609–614. PubMed Abstract
- 46.
Pfeiffer ML, DuPont HL, Ochoa TJ:
The patient presenting with acute dysentery—a systematic review.
J Infect.
2012; 64(4): 374–386. PubMed Abstract
| Publisher Full Text
- 47.
Khan AM, Faruque AS, Hossain MS, et al.:
Plesiomonas shigelloides-associated diarrhoea in Bangladeshi children: a hospital-based surveillance study.
J Trop Pediatr.
2004; 50(6): 354–356. PubMed Abstract
| Publisher Full Text
- 48.
Chan SS, Ng KC, Lyon DJ, et al.:
Acute bacterial gastroenteritis: a study of adult patients with positive stool cultures treated in the emergency department.
Emerg Med J.
2003; 20(4): 335–338. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 49.
Kavitha K, Latha R, Udayashankar C, et al.:
Three cases of Arcanobacterium pyogenes-associated soft tissue infection.
J Med Microbiol.
2010; 59(Pt 6): 736–739. PubMed Abstract
| Publisher Full Text
- 50.
Ferris MJ, Masztal A, Aldridge KE, et al.:
Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis.
BMC Infect Dis.
2004; 4(5): 1–8. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 51.
Chu YW, Wong CH, Chu MY, et al.:
Varibaculum cambriense infections in Hong Kong, China, 2006.
Emerg Infect Dis.
2009; 15(7): 1137–1139. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 52.
Flynn TR, Paster BJ, Stokes LN, et al.:
Molecular methods for diagnosis of odontogenic infections.
J Oral Maxillo fac Surg.
2012; 70(8): 1854–1859. PubMed Abstract
| Publisher Full Text
- 53.
Angelakis E, Roux V, Raoult D, et al.:
Human case of Atopobium rimae bacteremia.
Emerg Infect Dis.
2009; 15(2): 354–355. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 54.
Su CP, Huang PY, Yang CC, et al.:
Fusobacterium bacteremia: clinical significance and outcomes.
J Microbiol Immunol Infect.
2009; 42(4): 336–342. PubMed Abstract
- 55.
Auzias A, Bollet C, Ayari R, et al.:
Corynebacterium freneyi bacteremia.
J Clin Microbiol.
2003; 41(6): 2777–2778. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 56.
Lun ZR, Wang QP, Chen XG, et al.:
Streptococcus suis: an emerging zoonotic pathogen.
Lancet Infect Dis.
2007; 7(3): 201–209. PubMed Abstract
| Publisher Full Text
- 57.
Hillier DI, Ballal MS, Sanger R, et al.:
Veillonella dispar and Streptococcus bovis septic arthritis in a native shoulder joint.
Shoulder & Elbow.
2011; 3(4): 219–221. Publisher Full Text
- 58.
Griffen AL, Becker MR, Lyons SR, et al.:
Prevalence of Porphyromonas gingivalis and periodontal health status.
J Clin Microbiol.
1998; 36(11): 3239–3242. PubMed Abstract
| Free Full Text
- 59.
Grattarola C, Bellino C, Tursi M, et al.:
Helcococcus kunzii isolated from a sow with purulent urocystitis.
J Clin Microbiol.
2010; 48(8): 3019–3020. PubMed Abstract
| Publisher Full Text
| Free Full Text


Comments on this article Comments (0)