Introduction
Multifractals and fractals are related techniques mainly used in physics to characterize the scaling behavior of a system; they differ in that fractals look at the geometry of presence/absence patterns, while multifractals look at the arrangement of quantities such as population densities or biomass1. Scaling laws are an emergent general feature of ecological systems, and there is no a priori reason that power laws apply to ecological communities. If they do apply, they reflect constraints in their organization that can provide clues about the underlying mechanisms2,3.
Multifractals require that the object under study should be statistically self-similar, which means that a power-law could be fitted to data in a range of scales. But that does not mean that the power-law must be the best possible model. We can analyze the data without claiming that it is an exact multifractal. One of the advantages of multifractals is that they require fewer conditions on data than more classical statistics such as autocorrelation and variograms. These usually require isotropy and stationarity4 but multifractals can be used with anisotropic data5 and are inherently non-stationary6,7. Anisotropy and non-stationarity are often seen in spatial ecological distributions8.
Multifractals are associated with systems governed by random multiplicative processes9. In ecological systems, these processes can be given as the interaction of survival probabilities and compound growth10. Moreover, the presence of multiplicative process is argued to produce the log-normal-like shape of species-abundance distributions11. Also, random processes with spatial correlations can generate multifractals9; these kind of processes are part of neutral community models12,13 and are observed in natural communities14. Thus there are a priori reasons to think that multifractals can be applied to spatial ecological data. Indeed, they have been applied to vegetal communities15, tropical forest16, microphytobentos and periphyton biomass patterns1,17, and to the characterization of species-area relationships18–20.
Rank-abundance distributions are a representation of species-abundance distributions (SAD) that are a classical description of communities21. These have been used to compare different communities and to compare models and data, but different mechanisms can produce nearly identical SADs22. SADs are often presented using rank-abundance diagrams (RAD) where the log-abundance is plotted on the y-axis vs. rank on the x-axis21. RADs are equivalent to cumulative distributions23 and thus are a robust way to visualize the SAD without losing information24. If the rank of each species is incorporated in its spatial distribution, it forms a surface: the species-rank surface (SRS). This SRS can be analyzed and compared using multifractals.
But the application of multifractals is not widespread25; one of the reasons being the difficulty of using the available software for quantitative multifractal analysis (MFA). Here I present an open source software package for the application of multifractals, which can be integrated with R statistical software26.
Multifractal analysis
Several good introductions to multifractal methods applied to ecology are available15,27; thus I will only give a brief overview. Multifractals analyze the scaling properties of quantities distributed in a space that we assume to be two dimensional (a plane), but MFA can be used with one dimensional (time series) or three dimensional data28. A classical way to characterize multifractals is using the generalized dimensions Dq29, also called Renyi dimensions30. Dq has been used to characterize the probabilistic structure of attractors derived from dynamical systems5.
Another way to characterize multifractals is using the so called spectrum of singularities. This spectrum describes multifractals as interwoven sets each one with a singularity exponent α and a fractal dimension f (α)31. The two multifractal representations are equivalent, they display the same information in a different format. But with the spectrum of singularities, two quantities are estimated (α & f (α)) from data and are obtained with error. Instead, with generalized dimension only one quantity is estimated Dq, thus this method is preferred for statistical comparisons.
Estimation
To estimate multifractal spectra I used the method of moments based on box-counting32. I estimate generalized dimensions and the spectrum of singularities at the same time using the canonical method31. Here I describe only the Dq estimation; the steps for α and f (α) estimation are identical (only the formulae to calculate the quantities are different and can be found in the appendix of Saravia et al. (2012)1).
The spatial distribution that we are analyzing is covered with a grid, which is divided into N (∈) squares of side ∈. The contents of each square is called μi(∈). Then the so called partition function is computed as:
Where q is called moment order. The operation is performed for different values of ∈ and q, within a predetermined range. The generalized dimension is calculated as:
When q = 1, the denominator of the first term in Dq is undefined, so it must be replaced by the following expression:
In practical cases, as the limit can not be assessed, the dimensions are estimated as the slope of log(Zq) versus log(∈) in equation (1) (Figure 1). This is done for different q, provided that it is a real number which yields a graph of Dq in terms of q, called the spectrum of generalized dimensions (Figure 2).
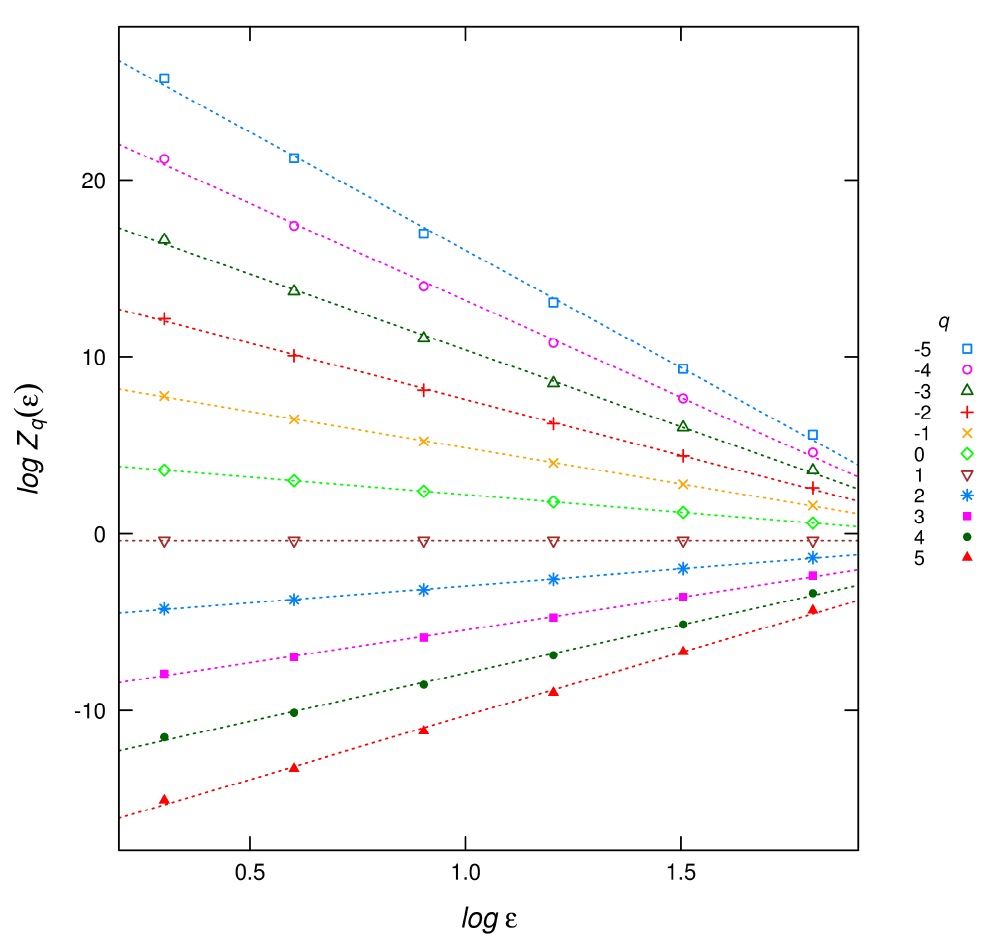
Figure 1. Plot of the linear regressions for different q used to estimate the Dq multifractal spectrum.
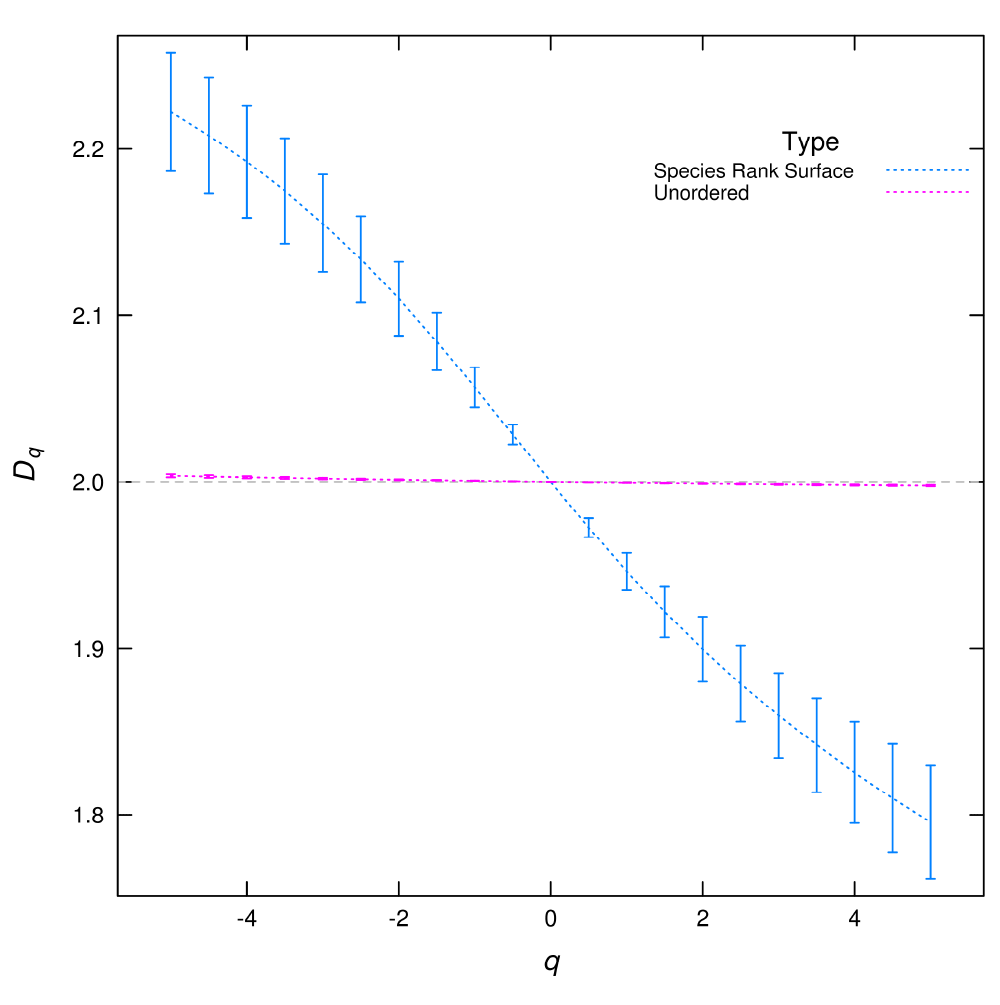
Figure 2. Dq multifractal spectrum calculated from species spatial distributions.
If each species is assigned a number at random (unordered) the Dq is almost flat corresponding to a uniform plus random noise distribution. But when the species rank surface (SRS) is used the Dq spectrum have a wide range of values. The error bars are the standard deviation obtained from the linear regressions used to estimate Dq.
To be an approximate multifractal, the relationship log(Zq) versus log(∈) should be well described by a linear relationship, although a linear relationship with superimposed oscillations is also acceptable27. A range of q and ∈ is fixed and then Dq is estimated using linear regressions. The coefficient of determination (R2) can be used as a descriptive measure of goodness of fit18.
Use of mfSBA software
The software was built and tested under Ubuntu 12.04 LTS Linux environment, using the GNU C++ compiler (v4.6.3). It requires the libtiff library for reading tiff images. It can be compiled under Windows environments using the GNU compiler and utilities for that operative system, but it was not tested.
You can download or clone mfSBA from https://github.com/lsaravia/mfsba (using git clone) and build it using the make utility
make -f mfSBA.mak
You can run it from the command line using the following command structure:
mfSBA inputFile qFile minBox maxBox numBoxSizes option
the parameters are:
inputFile: this file can have only two formats: 1) one-layer tiff 2) “sed” file format. Sed is an ascii format I invented to use with my own stochastic cellular automata models to represent a square grid of values. It has a header of two lines: the first line describes the two dimensions X Y of the data, and the second line describes the type of data. For this program, the type must be BI, which means that the values stored in the grid are real numbers with double precision. See the example file with a “.sed” extension.
qFile: this is a sed file with a vector of values representing the q’s used to calculate the multifractal spectrum.
minBox,maxBox,numBoxSizes: Minimum box size, maximum box size and maximum number of box sizes. The program uses box sizes in powers of two: if maxBox is greater than half of the image size, it is set to that value. If the number of boxes between minBox and maxBox is greater than numBoxSizes, the latter number of boxes will be used, discarding the biggest ones.
option: is an upper case character with four possibilities: N,S,D,A.
– N: analyze the input file as is.
– S: normalize the input file then analyze it. Normalization is done summing all the pixels values and dividing each pixel by that total. After that the sum of all values is one.
– D: add 1 to all the pixels then normalize as in S.
– A: normalize as in S and save the normalized image as a sed file.
Examples of input files are included with the source code, thus after compiling you could run the following command assuming a linux system:
./mfSBA b4-991008bio.sed q21.sed 2 256 20 S
Output
The program generates four output files, attaching a prefix to the original input file name:
t.inputFile: this file has a header line with field names and q + 2 columns. The first two columns are the box sizes and log box sizes used in the estimation. After that, each column corresponds to log (Zq (∈)) of equation (2) with the q’s specified in the qFile. This file could be used to visually check the linearity assumptions to calculate Dq as in Figure 1.
a.inputFile & f.inputFile: are similar to the previous file, but used to calculate α and f (α). The formulae are described in the appendix of Saravia et al. (2012)1.
s.inputFile: this file has a header line with field names and 10 columns. The first column is q. The second column is called Tau and is the result of the regression to calculate the limit in equation 2. Thus to obtain Dq we have to divide it by q − 1, except in the case of q = 1 that we take the value of the next column to get Dq. The third column is the value of α and the forth column f (α). After that, columns are the corresponding coefficients of determination R2 and standard deviations.
Species rank surface
I propose to extend the analysis of SAD attaching the rank of each species to its spatial distribution. In this way, the multivariate spatial distribution of all species can be summarized into a univariate distribution. I called this spatial distribution the species-rank surface (SRS), and it can be analyzed and compared using MFA. To construct the SRS, I first calculate the rank-ordering of the species by their abundance from biggest to smallest, starting from one. Then the rank is assigned to the spatial position of the individuals of each species, forming a surface. This landscape has valleys formed by the most abundant species and peaks determined by the rarest species, and the standard MFA can be applied. The program used to calculate this is called multiSpeciesSBA, and is included with the mfSBA source code. You can compile it using the following command:
make -f multiSpeciesSBA.mak
Then all the input files and parameters are identical to mfSBA except that the program expects an inputFile containing a multispecies distribution. So the inputFile should be composed of integer numbers each one representing one species. An example of a sed file with a multispecies spatial distribution is given in t64-0100.sed, this file was obtained using a spatially explicit neutral model with 64 species (available at https://github.com/lsaravia/neutral). You can use the following command to perform the MFA:
./multiSpeciesSBA t64-0100.sed q21.sed 2 128 20 N
R integration
Included with the source is a set of functions as an example to integrate the mfSBA software with the R language. You can load the functions inside R with:
source('Fun_MFA.r')
and then run the same given examples:
dq1<− calcDq_mfSBA("b4-991008bio.sed","q21.sed 2 256 20 S")
An interesting example is to compare the Dq
from the example multispecies spatial distribution untransformed
dq1<− calcDq_mfSBA("t64-0100.sed","q21.sed 2 512 20 S",T)
dq1$Site <− "Untransformed"
with the Dq from SRS
dq<− calcDq_multiSBA("t64-0100.sed","q21.sed 2 512 20 S",T)
dq$Site <− "Species Rank Surface"
dq <− rbind(dq,dq1)
and plot Dq with
plot_DqCI(dq)
In this plot (Figure 2), we can see that the rank ordering of the species in SRS is crucial to obtain a meaningful result. The Dq calculated from the unordered distribution is nearly flat, this corresponds to an almost constant spatial distribution with uncorrelated random noise.
The plot of the t.inputFile (Figure 1) gives a visual check of the regressions to obtain Dq:
plotDqFit("t.t64-0100.sed","q21.sed")
additionally the R2 values could be easily checked:
hist(dq1$R.Dq)
All the examples and more graphics are included in the file testMFA.r.
Conclusion
The multifractal spectrum can be used to describe spatial patterns of biomass, density, height, or any continuous variable. The mfSBA software is especially useful for remote sensing data because it can be used with tiff images. Multifractal patterns could be produced by the existence of multiplicative interaction between species and by spatially correlated random processes such as dispersal and growth9. Plant and animal species are generally aggregated in space thus is very likely that multifractal analysis can be used in a wide range of cases.
The analysis of SRS using Dq adds a new dimension to the comparison of species spatial distributions, because it can be used to compare spatial distributions of all species at the same time and also the abundances are accounted. An exploration of the results of different spatial patterns should be needed as a continuation of the present work.
The software presented here is oriented to obtain multifractal spectra for comparisons, rather than to obtain the true value. While the estimation methods used in mfSBA could be improved27,33, it has been used without trouble with the kind of data obtained in ecological studies1,34.
Multifractals can be successfully used to analyze several aspects of community spatial structure. With the advent of the big data era in ecology35 and the use of new technology to acquire spatial data36, new methods to analyze complex data sets are needed and multifractals could be an interesting addition to the ecologist’s toolbox.
Software availability
Zenodo: Multifractal estimation using a standard box-counting algorithm, doi: 10.5281/zenodo.765937
GitHub: Multifractal estimation using a standard box counting algorithm, https://github.com/lsaravia/mfsba
Competing interests
No competing interests were disclosed.
Grant information
The author(s) declared that no grants were involved in supporting this work.
Acknowledgments
I am grateful to Fernando R. Momo for our great conversations about ecological theory and David J. Harris for his language revision.
Faculty Opinions recommendedReferences
- 1.
Saravia LA, Giorgi A, Momo F:
Multifractal growth in periphyton communities.
Oikos.
2012; 121(11): 1810–1820. Publisher Full Text
- 2.
Brown JH, Gupta VK, Li BL, et al.:
The fractal nature of nature: power laws, ecological complexity and biodiversity.
Philos Trans R Soc Lond B Biol Sci.
2002; B 357(1421): 619–626. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 3.
Solé RV, Bascompte J:
Self-organization in complex ecosystems. Princeton University Press. 2006. Reference Source
- 4.
Fortin MJ, Dale MRT, ver Hoef J:
Spatial Analysis in Ecology.
Encyclopedia of Environmetrics.
John Wiley & Sons, Ltd. 2006. Publisher Full Text
- 5.
Harte D:
Multifractals: Theory and Applications. Chapman & Hall/CRC. 2001; 264. Reference Source
- 6.
Laurie H, Perrier E:
A multifractal model for spatial variation in species richness.
Ecological Complexity.
2010; 7(1): 32–35. Publisher Full Text
- 7.
Bez N, Bertrand S:
The duality of fractals: roughness and self-similarity.
Theoretical Ecology.
2011; 4(3): 371–383. Publisher Full Text
- 8.
Plotkin JB, Chave J, Ashton PS:
Cluster analysis of spatial patterns in Malaysian tree species.
Am Nat.
2002; 160(5): 629–644. PubMed Abstract
| Publisher Full Text
- 9.
Stanley HE, Meakin P:
Multifractal phenomena in physics and chemistry.
Nature.
1988; 335: 405–409. Publisher Full Text
- 10.
McGill BJ:
Strong and weak tests of macroecological theory.
Oikos.
2003; 102(3): 679–685. Publisher Full Text
- 11.
May RM, Crawley MJ, Sugihara G:
Communities: patterns. In: May RM McLean AR editors. Theoretical ecology: principles and applications. New York: Oxford University Press. 2007; 111–131. Reference Source
- 12.
Houchmandzadeh B, Vallade M:
Clustering in neutral ecology.
Phys Rev E Stat Nonlin Soft Matter Phys.
2003; 68(6 pt 1): 1–7. PubMed Abstract
| Publisher Full Text
- 13.
Yakimov BN, Iudin DI, Solntsev LA, et al.:
Multifractal analysis of neutral community spatial structure.
J Theor Biol.
2014; 343: 44–53. Publisher Full Text
- 14.
Condit R, Ashton PS, Baker P, et al.:
Spatial patterns in the distribution of tropical tree species.
Science.
2000; 288(5470): 1414–1418. PubMed Abstract
| Publisher Full Text
- 15.
Scheuring I, Riedi RH:
Application of multifractals to the analysis of vegetation pattern.
Journal of Vegetation Science.
1994; 5(4): 489–469. Publisher Full Text
- 16.
Manrubia SC, Solé RV:
Self-organized criticality in rainforest dynamics.
Chaos, Solitons & Fractals.
1996; 7(4): 523–541. Publisher Full Text
- 17.
Seuront L, Spilmont N:
Self-organized criticality in intertidal microphytobenthos patch patterns.
Physica A.
2002; 313(3–4): 513–539. Publisher Full Text
- 18.
Borda-de-Água L, Hubbell SP, McAllister M:
Species-area curves, diversity indices, and species abundance distributions: a multifractal analysis.
Am Nat.
2002; 159(2): 138–155. PubMed Abstract
| Publisher Full Text
- 19.
Yakimov BN, Bossuyt B, Iudin DI, et al.:
Multifractal diversity-area relationship at small scales in dune slack plant communities.
Oikos.
2008; 117(1): 33–39. Publisher Full Text
- 20.
Laurie H, Perrier E:
Beyond species area curves: application of a scale-free measure for spatial variability of species richness.
Oikos.
2011; 120(7): 966–978. Publisher Full Text
- 21.
McGill BJ, Etienne RS, Gray JS, et al.:
Species abundance distributions: moving beyond single prediction theories to integration within an ecological framework.
Ecol Lett.
2007; 10(10): 995–1015. Publisher Full Text
- 22.
Chave J, Muller-Landau HC, Levin SA:
Comparing classical community models: theoretical consequences for patterns of diversity.
Am Nat.
2002; 159(1): 1–23. PubMed Abstract
| Publisher Full Text
- 23.
Newman MEJ:
Power laws, Pareto distributions and Zipf’s law.
Contemporary Physics.
2005; 46(5): 323–351. Publisher Full Text
- 24.
Li L, Alderson D, Doyle JC, et al.:
Towards a Theory of Scale-Free Graphs: Definition, Properties, and Implications.
Internet Math.
2005; 2(4): 431–523. Publisher Full Text
- 25.
Seuront L:
Fractals and Multifractals in Ecology and Aquatic Science.
Taylor & Francis.
2009. Reference Source
- 26.
R Core Team: R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. 2013. Reference Source
- 27.
Borda-de-Água L, Hubbell SP, He F:
Scaling biodiversity under neutrality.
Scaling biodiversity.
2007; 347–375. Reference Source
- 28.
Lopes R, Betrouni N:
Fractal and multifractal analysis: a review.
Med Image Anal.
2009; 13(4): 634–649. PubMed Abstract
| Publisher Full Text
- 29.
Grassberger P:
Generalized dimensions of strange attractors.
Phys Lett A.
1983; 97(6): 227–230. Publisher Full Text
- 30.
Renyi A:
Probability Theory. Amsterdam: North-Holland. 1970. Reference Source
- 31.
Chhabra AB, Jensen RV:
Direct determination of the f(alfa) singularity espectrum.
Phys Rev Lett.
1989; 62(9): 1327–1330.
- 32.
Halsey TC, Jensen MH, Kadanoff LP, et al.:
Fractal measures and their. singularities: The characterization of strange sets.
Phys Rev A.
1986; 33(2): 1141–1151. PubMed Abstract
| Publisher Full Text
- 33.
De Bartolo SG, Primavera L, Gaudio R, et al.:
Fixed-mass multifractal analysis of river networks and braided channels.
Phys Rev E Stat Nonlin Soft Matter Phys.
2006; 74(2 pt 2): 26101. PubMed Abstract
| Publisher Full Text
- 34.
Saravia LA, Giorgi A, Momo FR:
Multifractal Spatial Patterns and Diversity in an Ecological Succession.
PLoS One.
2012; 7: e34096. Publisher Full Text
- 35.
Hampton SE, Strasser CA, Tewksbury JJ, et al.:
Big data and the future of ecology.
Front Ecol Environ.
2013; 11: 156–162. Publisher Full Text
- 36.
Michener WK, Jones MB:
Ecoinformatics: supporting ecology as a data-intensive science.
Trends Ecol Evol.
2012; 27(2): 85–93. PubMed Abstract
| Publisher Full Text
- 37.
Saravia LA:
Multifractal estimation using a standard box-counting algorithm.
ZENODO.
2014. Data Source


1) I highly appreciate the Author’s intention to provide ecologists with a tool for empirical multifractal analysis. However I think that the form ... Continue reading Several comments on the paper and the software:
1) I highly appreciate the Author’s intention to provide ecologists with a tool for empirical multifractal analysis. However I think that the form of C++ program created for the use in Linux environment is unsuitable. Numerous Windows-users cannot apply it or even to test. The same is true for R-interface. It works only in Linux environment.
2) After formula (3) Author states that «dimensions are estimated as the slope of log(Zq) versus log(e)», whereas it applies to mass exponent tau(q) estimation. It should be stated that mass exponents are divided by (q-1) to get Dq.
3) Author proposes application of R2 as a criterion of multifractality. That is a very bad choice. In my experience R2 depends strongly on q. It is in no way a measure of linearity. Instead I recommend application of Akaike information criterion for linear and quadratic fits in double log space.
4) I think illustration of program performance with species rank surface is not a good idea. Species rank surface and its analysis is something unknown to ecologists. As I understood it is proposed here for the first time. Section with its description leaves too many questions. How to construct the surface for the case of co-occurring species (i.e. not when each individual has its own cell of space)? What kind of neutral model is used for illustration? What is the interpretation of the final spectra?
1) I highly appreciate the Author’s intention to provide ecologists with a tool for empirical multifractal analysis. However I think that the form of C++ program created for the use in Linux environment is unsuitable. Numerous Windows-users cannot apply it or even to test. The same is true for R-interface. It works only in Linux environment.
2) After formula (3) Author states that «dimensions are estimated as the slope of log(Zq) versus log(e)», whereas it applies to mass exponent tau(q) estimation. It should be stated that mass exponents are divided by (q-1) to get Dq.
3) Author proposes application of R2 as a criterion of multifractality. That is a very bad choice. In my experience R2 depends strongly on q. It is in no way a measure of linearity. Instead I recommend application of Akaike information criterion for linear and quadratic fits in double log space.
4) I think illustration of program performance with species rank surface is not a good idea. Species rank surface and its analysis is something unknown to ecologists. As I understood it is proposed here for the first time. Section with its description leaves too many questions. How to construct the surface for the case of co-occurring species (i.e. not when each individual has its own cell of space)? What kind of neutral model is used for illustration? What is the interpretation of the final spectra?