Introduction
In the last few years there has been a rapid growth of biological network data, including protein-protein interaction (PPI) networks, metabolic networks and regulatory networks. Among these, PPI networks are important in several biological phenomena such as signaling, transcriptional regulation and formation of multi-enzyme complexes1.
Comparing PPI networks of evolutionary distant species can help to understand some mechanisms underlying a specific function or process, which the sequence comparison alone cannot explain. Local network alignment aims to compare networks of different species, in order to find conserved protein complexes or pathways.
In literature, several network alignment algorithms have been described together with their implementations2–5, however none of them is fully integrated into Cytoscape.
Here, we describe a Cytoscape app implementing the GASOLINE algorithm for multiple local alignment of PPI networks. Aligned proteins can be associated with GO annotations for further functional analysis of alignments. To the authors knowledge, it is the first tool that online computes and visualizes local alignments in a user-friendly way, without requiring any post-processing operations.
Implementation
The GASOLINE app is based on the homonymous greedy and stochastic algorithms introduced in6.
The app has been written in Java version 7 and designed following a classic Model-View-Controller (MVC) model. The Model part is represented by the classes implementing the algorithm and the auxiliary data structures. The View part is composed by two Java Panels; one for setting all the input and output parameters, and one for listing local alignments and handling their visualization.
The Controller part ensures the communication between the Model and the View and is implemented by different Cytoscape Task classes, one for each process performed by GASOLINE (i.e. checking file format, computing alignments, importing networks, protein description and GO annotations, building alignment graphs). Each Task class properly notifies the corresponding view class when a task has been completed.
Input networks are imported as text files and then internally represented in two different ways, in order to optimize the performance of our algorithm. We used CyNetwork and CyNetworkView objects for network alignment visualization and custom classes for computing alignments. For all the imported networks, the corresponding Cytoscape view is initially disabled to reduce memory consumption.
The main component of GASOLINE is represented by a tabbed panel named “GASOLINE”, in the Control Panel of Cytoscape (Figure 1). Through the interface users can provide the following information:
“Similarity information”, to upload orthology similarity scores between proteins of different species
“Networks”, for selecting two or more networks to align
“Parameters setting”, to modify the default GASOLINE parameters
“Optional parameters setting”, for setting other advanced input parameters
“Ontologies”, to upload GO terms linked to the proteins of the aligned networks
“Output”, to specify the folder where the final alignments will be saved
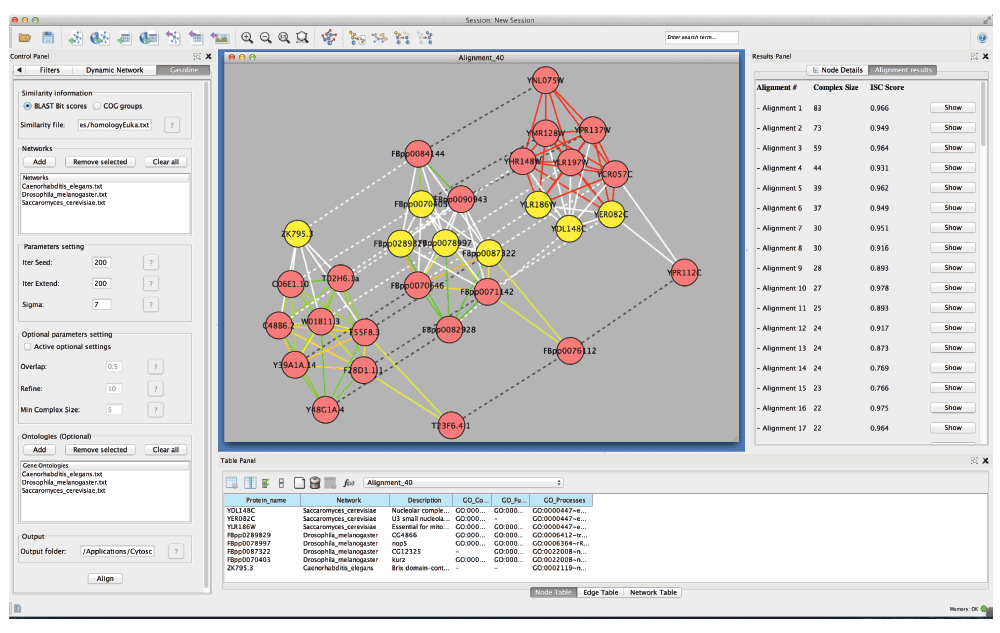
Figure 1. GASOLINE Cytoscape panel.
The button labeled “?”, when present, explains the meaning of a specific function or parameter of GASOLINE, whenever the mouse arrow hovers over that button.
In the following subsections, we will describe all the required steps to run GASOLINE on a set of PPI networks.
Loading input data
Before running GASOLINE, the user needs to upload input data, including:
a) Two or more networks to be aligned;
b) A file of orthology BLAST bit scores between proteins of different species;
c) A set of GO terms linked to the proteins of each network.
The GO terms file is not mandatory and can be omitted. Networks are given as a list of weighted edges and can be uploaded from the “Networks” panel.
Orthology data can be uploaded through the “Similarity information” panel. They can be supplied in two different formats: “BLAST Bit scores” or “COG groups”.
The “BLAST Bit scores” format is a text file where each row has a couple of proteins of different species followed by their corresponding BLAST bit score.
Files in the “COG groups” associate a list of orthology groups (e.g. KOG, NOG, COG groups) to the proteins of aligning networks. The “COG groups” format can be more convenient when aligning many networks since the all the possible pairwise bit scores are many.
GO categories can be optionally uploaded from “Ontologies” panel. They are provided as text files, where a list of GO cellular components, processes and functions is associated to each protein.
Whenever GO categories are provided as input, the list of GO terms for a specific protein is added as node attribute in Cytoscape, these are accessible from the “Node Browser” tabbed panel once GASOLINE ends the computation and the local alignments are ready to be visualized.
Setting the parameters
The main input GASOLINE parameters are specified in the “Parameters setting” panel. These include:
“Iter Seed”: the number of iterations of Gibbs sampling in the bootstrap phase
“Iter Extend”: the number of iterations of Gibbs sampling in each extension step of the iterative phase
“Sigma”: minimum network degree of nodes that can be selected as seeds in the initial phase
Values for “Iter Seed” and “Iter Extend” depend on the number of aligning networks: the more networks we have, the higher these values should be. However, based on the experiments performed on real PPI networks and reported in6, we empirically estabilshed that 200 iterations of Gibbs sampling in both phases are enough to produce reliable results for up to 25 networks. GASOLINE is very fast; it computes the results on 25 networks in a few minutes.
The choice of “Sigma” implies a tradeoff between speed and accuracy of GASOLINE: the higher the σ, the faster is GASOLINE but the lower its accuracy. If networks are very sparse (like most of the existing PPI networks), low values of sigma (1 or 2) are recommended.
The “Optional parameters setting” panel contains three more input parameters:
“Overlap”: a value between 0 and 1, denoting the maximum allowed fraction of common nodes between two alignments, in order to be considered distinct. If two alignments have many nodes in common, the one with the least number of nodes is discarded from the final set;
“Refine”: the number of iterations of the GASOLINE iterative phase
“Min Complex Size”: the minimum size of conserved complexes in the final set of local alignments
These parameters can be modified by checking the box “Active optional settings”, otherwise the default values will be used.
Note that a high value of the “Refine” parameter can be used to increase the accuracy of the local alignments, but the algorithm will be more time consuming. In our tests6, we experienced that a value of 10 guarantees the best trade-off between speed and accuracy of GASOLINE.
For the “Overlap” and “Min Complex Size” parameters, we suggest 0.5 and 5 as default values, respectively.
Finally, the user can specify an output folder for the final alignments, by clicking on the text field next to the “Output folder” label of the “Output” panel. Each local alignment will be stored in a separate text file inside the specified folder, containing the list of aligned sub-graphs and the one-to-one mapping between aligned nodes.
Running GASOLINE
Once all the required input files are provided and all the parameters are set up, GASOLINE can be executed by clicking on the “Align” button. Then, a task window will appear describing the progress of the algorithm.
Visualizing local alignments
When GASOLINE ends, a table containing all the computed local alignments is shown on the right side of the “Results panel” of Cytoscape (Figure 1). The table reports, for each alignment, the size of the aligned complexes and the ISC score.
Each row of the table contains a “Show” button, for the visualization of the corresponding alignment graph on the left side of the “Results Panel” of Cytoscape (Figure 1).
In the alignment graph, each node is labeled with the ID of the corresponding protein. If GO annotations have been provided, the user can select a node and view the description of the protein and its corresponding GO terms from the “Node Attribute Browser” tabbed panel of Cytoscape.
Two kinds of edges are shown:
Intra-edge, linking proteins of the same network, which are represented with solid colored lines
Inter-edge, linking proteins of different networks that map one another in the local alignment, which are drawn with dashed lines
Colors of intra-edges depend on the probability p of the corresponding protein-protein interaction: for low values of p colors range from green to yellow, for high values of p colors range from yellow to red. Weights are automatically associated to edges as attributes, so the user can select an edge and retrieve its weight from the “Edge Attribute Browser” tabbed panel of Cytoscape.
Layout “Kamada-Kawai” has been used for the visualization of the alignment graph.
Results
Finally, we show an example of the workflow, using three well known PPI networks C. elegans, D. melanogaster, S. cerevisiae) taken from the STRING database7, considering only experimentally validated interactions. We also annotated proteins by using a set of GO annotations and protein descriptions taken from BioDBnet8.
Following the steps described in the Implementation section, we loaded the three networks and ran GASOLINE, using default parameters (IterSeed = 200, IterExtend = 200, Sigma = 7, Overlap = 0.5, Refine = 10, MinComplexSize = 5).
GASOLINE took 110 seconds to complete the task and returned many known conserved complexes with a high degree of topological conservation (ISC between 80 and 90%). These include the large and small subunits of ribosomes (64 proteins), a serine/threonine kinase complex (34), the spliceosome (28), a DNA repair complex (24 proteins, Figure 2), the V-ATPase complex (17) and the ARP2/3 complex (16).
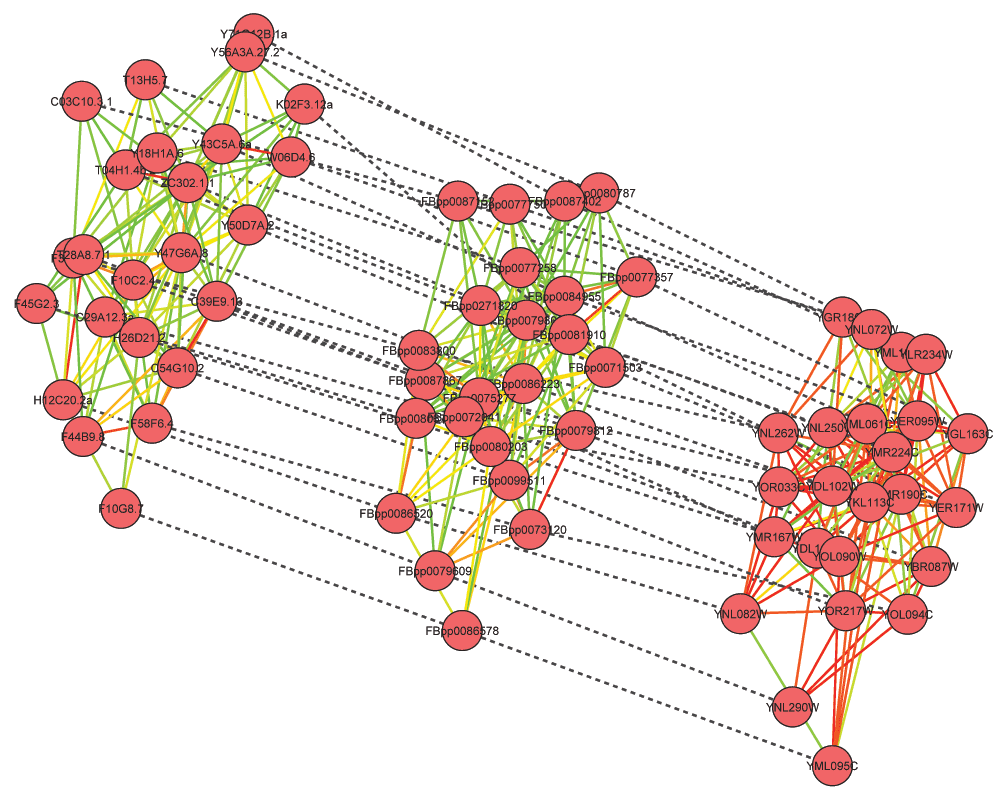
Figure 2. Alignment of DNA repair complex in C. elegans, D. melanogaster and S. cerevisiae.
Conclusions
In this paper, we presented GASOLINE, an app for Cytoscape 3 for computation and visualization of multiple local alignments of protein-protein interaction networks. To the best of our knowledge, it is the first Cytoscape plugin for computation and visualization of multiple local alignment of biological networks.
GASOLINE offers a user-friendly interface and an easy 2D visualization of local alignments. Moreover, alignments can be further investigated, by attaching GO terms to the proteins of aligning networks.
Software availability
The GASOLINE app, as well as datasets of real PPI networks and orthology files that can be directly used to run the algorithm, can be downloaded from the GASOLINE website: http://ferrolab.dmi.unict.it/gasoline/gasoline.html.
The GASOLINE plugin can also be downloaded from the Cytoscape App Store: http://apps.cytoscape.org/apps/gasoline.
On our website, there is also a complete documentation on the GASOLINE plugin, with more details about the format of input and output data, and a JAR file for running our algorithm in local with any platform.
Latest source code: https://github.com/GMicale/GASOLINE
Source code as at the time of publication: https://github.com/F1000Research/GASOLINE/releases/tag/V1.0
Archived source code as at the time of publication: http://www.dx.doi.org/10.5281/zenodo.104629


Comments on this article Comments (0)