Keywords
.bioinformatics, cytoscape, keggscape, data integration, pathway data
This article is included in the Cytoscape gateway.
This article is included in the Japan Institutional Gateway gateway.
.bioinformatics, cytoscape, keggscape, data integration, pathway data
Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg)1 is a widely used biological database of high-level biological functions. It contains pathway data sets that have comprehensive annotations and high quality human-curated, hand-drawn diagrams. Most biological pathway databases store data as machine-readable graph topologies, which leave much of the details about how the diagrams were drawn excluded from the data files. This is a problem when third-party developers want to reproduce the pathway diagrams in their applications. In contrast, the KEGG PATHWAY database stores graphics information in machine-readable KEGG Markup Language (KGML, http://www.kegg.jp/kegg/xml) format. Thus, in these pathway diagrams, biological entities, such as enzymes or compounds, are manually laid-out and the diagrams are easy to understand for biologists.
The KEGG PATHWAY database is deployed as a web application using static bitmap images for pathway diagrams, and user-provided date is integrated with KEGG Mapper (http://www.genome.jp/kegg/mapper.html). Furthermore, KEGG Atlas (http://www.genome.jp/kegg/atlas.html) provides a comprehensive network view of global metabolic pathways. Recent improvements to KEGG Atlas, such as Pathway Projector2 and iPath23, have made it possible to perform basic data integration and visualization like mapping the expression values to node graphics. However, despite these features, it is difficult to integrate external data sets and create custom visualization. Furthermore, they are limited to those on existing desktop pathway analysis applications. To ameliorate these problems, several projects for integrating a user’s own models onto the KEGG pathways have therefore been developed (CytoSEED Cytoscape app4, KEGGtranslator5).
Cytoscape6,7 is a de-facto standard software platform for biological network analysis and visualization. One of its advantages is its large collection of apps for a variety of biological problem domains, such as Gene Ontology term enrichment analysis (BiNGO8) and statistical network analysis (CentiScaPe9), which are also mostly open source software. Additionally, Cytoscape has a flexible network visualization function and is optimized for large-scale network analysis. There are several applications dedicated to biological pathway analysis (Vanted10, VisANT11) that support KGML by default. Although Cytoscape does not have a built-in function to load biological pathways, if this task is done with a separate app, users can take advantage of its large-scale network analysis features, variety of analysis apps, and data visualization of biologists-friendly human curated pathways.
The goal of our new Cytoscape app, KEGGscape, is to bridge the flexibility of fully-featured network analysis platforms with the high-quality pathway diagrams available in the KEGG PATHWAY web application. KEGGscape, a successor of KGMLReader (http://apps.cytoscape.org/apps/kgmlreader) for Cytoscape 2 series, is an app that imports KEGG pathway diagrams from KGML files and provides a new way to use KEGG pathway diagrams as data integration blueprints in cooperation with Cytoscape core features and an existing variety of apps. KEGGscape is completely re-designed for the new Cytoscape 3 API and supports signaling pathways in addition to metabolic pathways, including the global metabolic pathways used in KEGG Atlas (Figure 1). In this paper, we present a basic design and implementation of KEGGscape and an example workflow utilizing KGML files, and experimental data to create information-rich pathway visualizations for clarifying omics-scale data sets. KGMLReader is the first open-source Cytoscape app that reads the graphics details of KGML files, and KEGGscape was designed to use standard Cytoscape features only. These feature enable users to use KEGG pathways with other data sets easily.
KEGGscape is a Cytoscape 3 app written in Java programming language and is designed to load pathway data files in KGML format. KGML is an XML file format designed by the KEGG project and contains the topology of pathways and visual representations of all elements in the diagram. KGML has formal specification as a DTD (Document Type Definition) file, which enables the use of unmarshaller (https://jaxb.java.net) for converting XML elements directly into Java objects. This conversion creates two types of data: pathway topology and its graphical representations. Pathway topology and its properties are converted into CyNetwork and CyTable objects, which are the standard data model in Cytoscape 3. In KGML, all graphical information, such as the color of enzymes or shape of compounds is stored under <graphics> tag. Instead of setting the graphics details of nodes and edges directly from this information, Cytoscape generates Visual Style, which is a collection of default visual properties and visual mapping function, for each pathway based on the information under this tag. KEGGscape follows a standard CyNetworkReader design guideline, which enables Cytoscape to detect KGML files automatically.
Figure 2 shows an example of a pathway analysis workflow with KEGGscape. To take advantage of the flexible visualization and analysis features in Cytoscape, users need to import as much information as possible for the pathways they want to analyze. Although Cytoscape is a powerful tool for biological data integration, it is not the best platform for data preparation or cleansing. Users can instead prepare annotations and experimental data sets for the pathway using tools of their choice, such as R (Bioconductor12), Python, or Excel. Once the data files are ready, Cytoscape can read them into an on-memory session and visualize the data on the KEGG pathways. Imported data sets only use standard Cytoscape data objects, and users can then access all of the standard Cytoscape features to create custom pathway visualizations. An actual workflow will be presented in a later section.
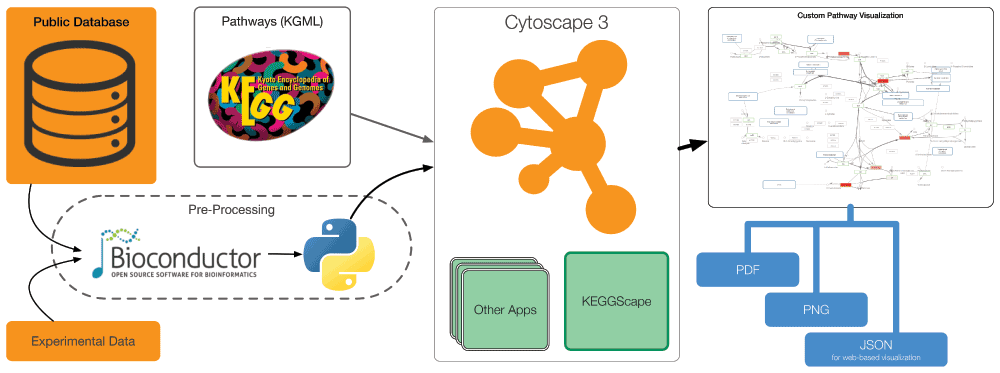
Cytoscape with KEGGscape can be used as a part of larger workflows to publish integrated pathway visualizations as vector graphics, bitmap images, or JSON for web-based visualization using Cytoscape.js (http://cytoscape.github.io/cytoscape.js/).
Although KEGGscape can read all information of the pathways saved in KGML files, some of the pathway visualizations in Cytoscape look slightly different from the original hand-drawn diagrams available on the KEGG website. The cause of this issue is missing graphics information in the KGML files. Figure 3 is a side-by-side comparison of the same pathway visualization (human MAPK signaling pathway; KEGG ID: hsa04010). The original diagram (left) contains several background visual annotations that are not visible in the visualization created by Cytoscape (right). The hand-drawn compartmental annotations are not encoded in KGML files, which means they cannot be reproduced by KEGGscape.
As an example workflow, we integrated and visualized a KEGG pathway and gene expression profile using KEGGscape and external tools. In this example, the differentially expressed genes between two groups, mutants and controls, in a global expression profile are mapped on the KEGG pathway, as too are the t-test results.
To perform this pathway analysis in Cytoscape, we used Bioconductor (http://www.bioconductor.org/) to prepare the gene expression matrix data. We normalized Affymetrix GeneChip data by the robust multi-array average (RMA) method with the Bioconductor packages ecoliLeucine13 and affy14. The leucine regulatory protein (Lrp) is a DNA binding protein and known as a leucine responsive global regulator15. The p-value for each probeset between four lrp mutant strains and four control chips was calculated by rowttest method in genefilter package16. From this calculation, we obtained a list of genes that are differentially expressed (p-value < 0.05). We sent these probeset identifiers to KEGG Mapper and picked the highest hit, which was the glycine, serine and thereonine metabolic pathway (KEGG ID: eco00260) for visualization.
To create a visualization using all the data sets, we imported the KGML file of eco00260 and the p-value matrix file prepared in the previous data preparation to Cytoscape 3, and merged the matrices with a custom Python script (Figure 4). Because Cytoscape does not support fuzzy key matching, we used our Python script to append a key column to the p-value matrix to utilize the Cytoscape table merge tool.
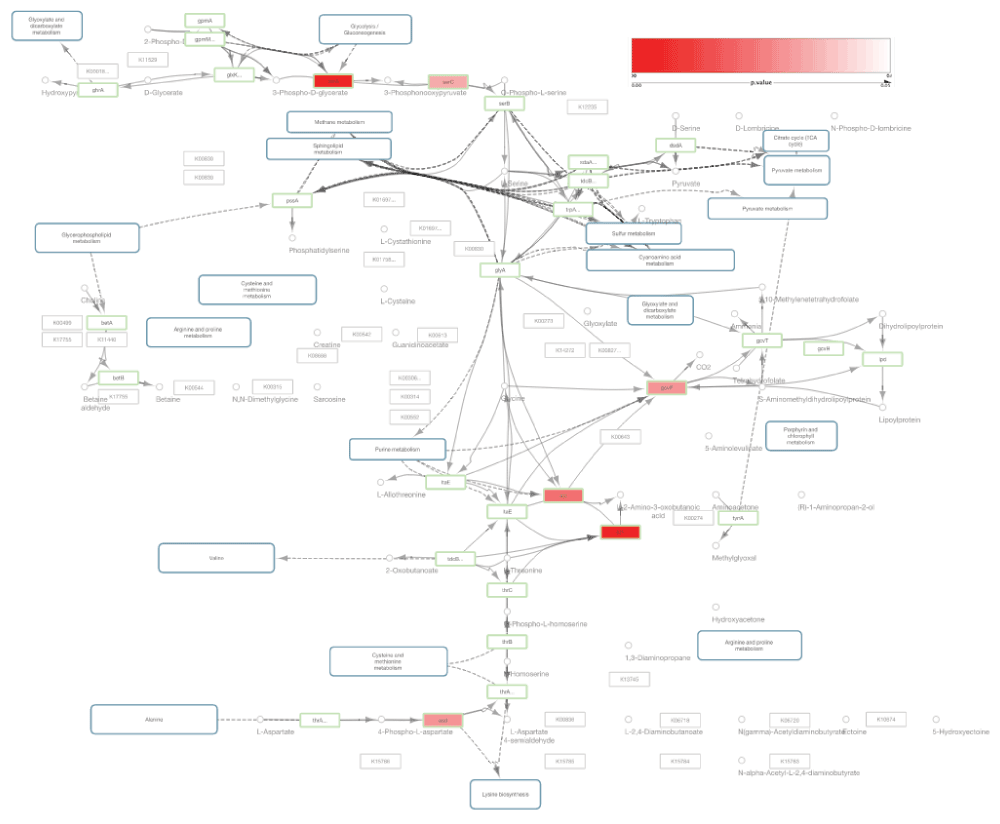
Green border nodes are KO (KEGG Orthology) annotated nodes. Red colored nodes include differentially expressed genes (p-value < 0.05).
The node table in Cytoscape for the imported KGML had KEGG gene annotations. The gene IDs for each enzyme node were used as keys for merging the KGML node table and p-value matrix. In this Figure 4, node colors in the original KEGG pathway were mapped to node border colors and p-values were mapped to node color gradient (red to white) to visualize the significantly expressed genes.
In this paper, we presented the design and implementation of KEGGscape and an example analysis workflow integrating global gene expression profiles and KEGG pathways using KEGGscape and two external tools, Bioconductor and Python. The workflow demonstrates how users can integrate omics data in an interactive pathway diagram.
Current workflow can map arbitrary omics data onto interactive KEGG pathway diagrams, but it requires some manual editing to create informative visualizations. To minimize the manual process in the workflow, we plan to implement a collection of utility Python scripts to manipulate networks and Visual Styles via RESTful API, which will be published as a part of the Cytoscape 3.2.0 release. This set of Python scripts works to merge pathway related table metadata (omics profiles, non-KEGG pathway metadata) from external platforms like R and Cytoscape to automate common tasks in the visualization process.
The app website: http://apps.cytoscape.org/apps/keggscape
Latest source code: https://github.com/idekerlab/KEGGscape
Source code as at the time of publication: https://github.com/F1000Research/KEGGscape/releases/tag/V1.0
Archived source code as at the time of publication: http://dx.doi.org/10.5281/zenodo.1056017
License: Apache License Version 2.0
SK guided the GeneChip data preparation and the statistical tests, KT guided the total system design of the project. KN designed and implemented the software under KO’s supervision and performed sample analysis and visualization. KO contributed to the writing of this article.
This work was supported by National Bioscience Database Center (NBDC) of the Japan Science and Technology Agency (JST).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would like to thank the Google Summer of Code program and Biohackathon attendees for helpful suggestions at the early stage of KEGGscape development, and Peter Karagiannis for reading this article and for useful comments.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
The paper describes an interesting and relevant Cytoscape plugin; it is well written (there are a few typos), but I think it can be improved in places. An important reservation is that I cannot determine at this moment how common or serious the data loss in KGML reading is (see below). That is it should be clarified whether this paper is a first report, a work in progress or a finished project..
Title and Abstract
I find the title appropriate, but it may be promising more than what the app actually does. As far as I can tell, the app is not a general solution, but one specific for KEGG. The abstract is short on setting the state of the domain and, like the Introduction, does not refer to existing research in fields beyond their own. In this way, the abstract is less than clear what unsolved problem was resolved.
Introduction
The Introduction sufficiently explains the setting of the plugin. One thing that strikes me is the lack of references to related work. For example, it mentions "most biological pathway databases" without citing or even naming them, which I find suboptimal as they critique those databases and to which the authors contrast their solution. Unlike the Introduction currently seems to suggest, KGML is not the only pathway format that tracks graphical information (e.g. GPML used by PathVisio). The first paragraph currently suggests it solves a problem that others have been tackling too. (P.S. under what license is the KGML specification available?).
The Introduction focuses on technical issues of the integration. It could also say a bit about identifier mapping and the role of licensing in data integration. That is, given the enormous impact of KEGG it can be wondered why this kind of integration with KEGG has not been done yet, because people have done this for other data sets for years (both open source and proprietary). Or, if it has, it should be cited.
Minor comments:
Figures
The figures are too hard to read, both in print (where it is impossible) but also in the PDF. This should really be addressed.
Implementation
The Implementation sections can be improved: the section is short on details on the KEGGscape source code, the design of the app and the KGML reader in particular, build instructions, development model, testing (tests seem absent in the code repository).
I also note issues with the KGML readers. For example, connections clear on the KEGG website are missing and misplaced when read into Cytoscape (e.g. between glycine and sarcosine, and the link between Purine metabolism, see Fig. 4) and labels are often misleadingly placed and often unconnected (a limitation of KEGG), though accurately copied from the KGML, it seems. The first data-loss clearly indicates a problem with the reader. These things must be discussed in the Limitations subsection, in my opinion (and/or returned to in the Future Plan at the end).
Minor comments:
Results
The Results section seems to accurately describe what they did, but not how they did this. I think something like a methods section (or an Open Notebook) that explains how the steps are performed (in more detail) would be helpful. However, in the end I was able to figure out how to import a KGML file (which is not under Import).
Another example is reference to a custom Python script without description. In fact, thinking about this again, for an app that does data integration, I would actually expect the app to take care of data processing as much as possible. While I understand that general preparation starting from raw data is best done in other tools, this Python script does not seem to do more than format handling of some kind. Is that correct? If so, why is this not done by the app? Fortunately, this observation was made by the authors too, and they return to it in the Future plan subsection.
Minor comments:
Conclusion
The Conclusion section does not sufficiently summarize the paper: it should reiterate what the problem is that is solved and what makes KEGGscape unique. Although I do not want to imply that I think this paper is not relevant (it is)!
Also the limitations should be returned to here (those already listed in the paper, and those missing), particularly in relation to the "Future plan". The authors’ focus should really be on getting the KGML reading performing well: data loss, and data corruption are serious issues. A decent testing toolkit as Maven supports, may be a good start here, but is currently missing from the source code repository.
Minor comments:
Competing Interests: I am contributing to the open source WikiPathways, BridgeDb, and PathVisio projects. I have no competing financial interest.
KEGGscape is valuable Cytoscape app that enables flexible visualization and manipulation of KEGG pathway maps. The capability for visualizing data sets (e.g., gene expression levels) on pathways in Cytoscape is particularly appealing. However, the example presented in the paper does not contain sufficient information to enable a reader to duplicate the process. First, the authors do not cite the origin of the gene expression dataset they use in their example. Second, as far as I can tell, there is no tutorial provided with KEGGscape. At a minimum, the authors should provide instructions for opening a KGML file and loading a dataset.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 01 Jul 14 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)