Keywords
immunohistochemistry, antibody, ARID1A, tissue
This article is included in the Antibody Validations gateway.
immunohistochemistry, antibody, ARID1A, tissue
ARID1A (AT-rich interactive domain 1a) is a member of the SWI/SNF family and its loss has been implicated as a factor in multiple premalignant and malignant conditions, including Barrett’s oesophagus and oesophageal carcinoma as well as endometrial and clear cell ovarian carcinomas and their precursor endometriotic lesions1–4. The ARID1A antibody from Human Protein Atlas is a rabbit antibody generated against a PrEST (Protein Epitope Signature Tag) fragment of the ARID1A gene and affinity purified against the same fragment5. It is thus designated as being “mono-specific” in that the affinity purification removes any non-specific or low affinity binders to the peptide. Through the Human Protein Atlas, the antibody has been tested on a wide variety of human tissue types and human malignancies, as well as for expression in immunofluorescence on U-2 OS, A-431 and U-251 MG cell lines. This demonstrates a nuclear expression in all cell lines and in the majority of tissue types6. However, the Western blot data were not supportive and did not produce staining corresponding to the expected size, although the data from a protein array did confirm a peak at the expected size. The antibody has been used to stain human colorectal cancers7, clear cell carcinomas8 and on a variety of clear cell cancer cell lines9 by immunohistochemistry.
To our knowledge, whilst the sequence homology between mouse and human ARID1A is 95%, this antibody has not been qualified using knockout tissue and has not been tested or published on murine tissue and this work represents the first data to do so.
Details of all reagents with reference to the immunohistochemical staining procedure can be found in Table 1.
Anti-ARID1A is a monospecific rabbit polyclonal generated to a PrEST sequence – PGLGNVAMGPRQHYPYGGPYDRVRTEPGIGPEGNMSTGAPQPNLMPSNPDSGMYSPSRYPPQQQQQQQQRHDSYGNQFSTQGTPSGSPFPSQQTTMYQQQQQNYK (Table 2). The lot number used for all validations was A40072 and for subsequent staining was D81856. A concentration of 1 µg/ml was used for initial validations and 0.5 µg/ml for final runs.
Donkey anti-rabbit biotin (Jackson Immunoresearch, Table 2) is specific for Rabbit IgG (Heavy and Light chains) and was affinity purified to remove cross-reactions to Bovine, Chicken, Goat, Guinea Pig, Syrian Hamster, Horse, Human, Mouse, Rat and Sheep. All slides were stained with a concentration of 4.8 µg/ml.
Anti-GAPDH was used as a loading control for Western blots and was a rabbit monoclonal (Cell Signaling, Table 2). Detection antibody for the Western blot for ARID1a was Goat anti-rabbit IR Dye 680LT (Li-Cor Biosciences, Table 2) used at a concentration of 0.1 µg/ml and detection of GAPDH was Goat anti-rabbit IR Dye 800CW (Li-Cor Biosciences, Table 2).
All tissues and cell pellets used during the validation were fixed in Neutral Buffered Formaldehyde as specified (Table 3) before being transferred directly to 70% ethanol for no longer than 3 days. Tissue processing was conducted on a Leica ASP300 through graded ethanols before clearing in xylene and impregnation in molten paraffin wax (Fisher). All tissue sections were cut on a Leica rotary microtome at 3 µm.
| Species | Tissue Type | Strain/Cell line | Details | Fixation Time |
|---|---|---|---|---|
| Murine | Uterus | C57Bl6 | Female | 16 hrs |
| Murine | Uterus | C57Bl6 KO Arid1afl/fl | Female | 24 hrs |
| Human | Cell Pellet | ES-2 | 20 hrs | |
| Human | Cell Pellet | RMG-II | 20 hrs |
Western blot. Protein was extracted from the two clear cell carcinoma cell lines using a Tris-EDTA lysis buffer and run on a non-denaturing 3–8% Tris-acetate gel (Life Technologies). Following electrophoresis, the transfer membrane was probed with 0.2 µg/ml of anti-rabbit ARID1A (HPA005456) at 4°C overnight and 0.1 µg/ml anti-GAPDH (14C10) for the same length of time. Detection of the anti-rabbit ARID1A was with Goat anti-rabbit IRDye 680LT (Li-Cor Biosciences) and the GAPDH was with Goat anti-rabbit IRDye 800CW (Li-Cor Biosciences) both at 0.1 µg/ml.
Immunohistochemistry. Slides were deparaffinised and rehydrated on a Leica ST5020 using Xylene (Sigma) for 2 × 10 mins and ethanol (Fisher), 2 × 100% ethanol followed by 1 × 70% ethanol for 5 mins each. Following staining, all slides were dehydrated, cleared and mounted and coverslipped in DPX (Fisher).
The antibody was validated on a Leica BondMax instrument using a Leica Intense R kit to a standardised in-house protocol. All reagents were from Leica as part of the Intense R kit and were conducted at room temperature, unless otherwise specified. All staining steps included individual washes in Leica Bond Wash after each step, as part of the protocol (Table 4). A full protocol for the validated conditions can be found in the supplementary material. In this protocol, the step named “primary” refers to the anti-ARID1a primary antibody.
A slide using the same conditions and retrieval but omitting the primary antibody was used to control for any background staining due to the retrieval and detection steps.
Imaging. All slides were digitised using a Leica Scanscope AT2 at 0.5 µm/pixel resolution. Datasets can be viewed by downloading the Leica Imagescope free viewer at http://www.leicabiosystems.com/pathology-imaging/aperio-epathology/integrate/imagescope/.
To determine the correct cell line to utilise and to confirm the equivocal Western blot data from Human Protein Atlas, the antibody was used to stain a Western blot of two cell lines; ES-2 and RMG-II, both of which are cell lines derived from clear cell carcinoma. It could be demonstrated that the HPA ARID1A antibody showed positive expression in ES-2 cell lines at the expected size of 270 kDa and no staining for RMG-II. The loading control of GAPDH showed that there were no loading issues (Figure 1). Thus, these cell lines were chosen to be grown, formalin fixed and processed into paraffin wax for immunohistochemical validation.
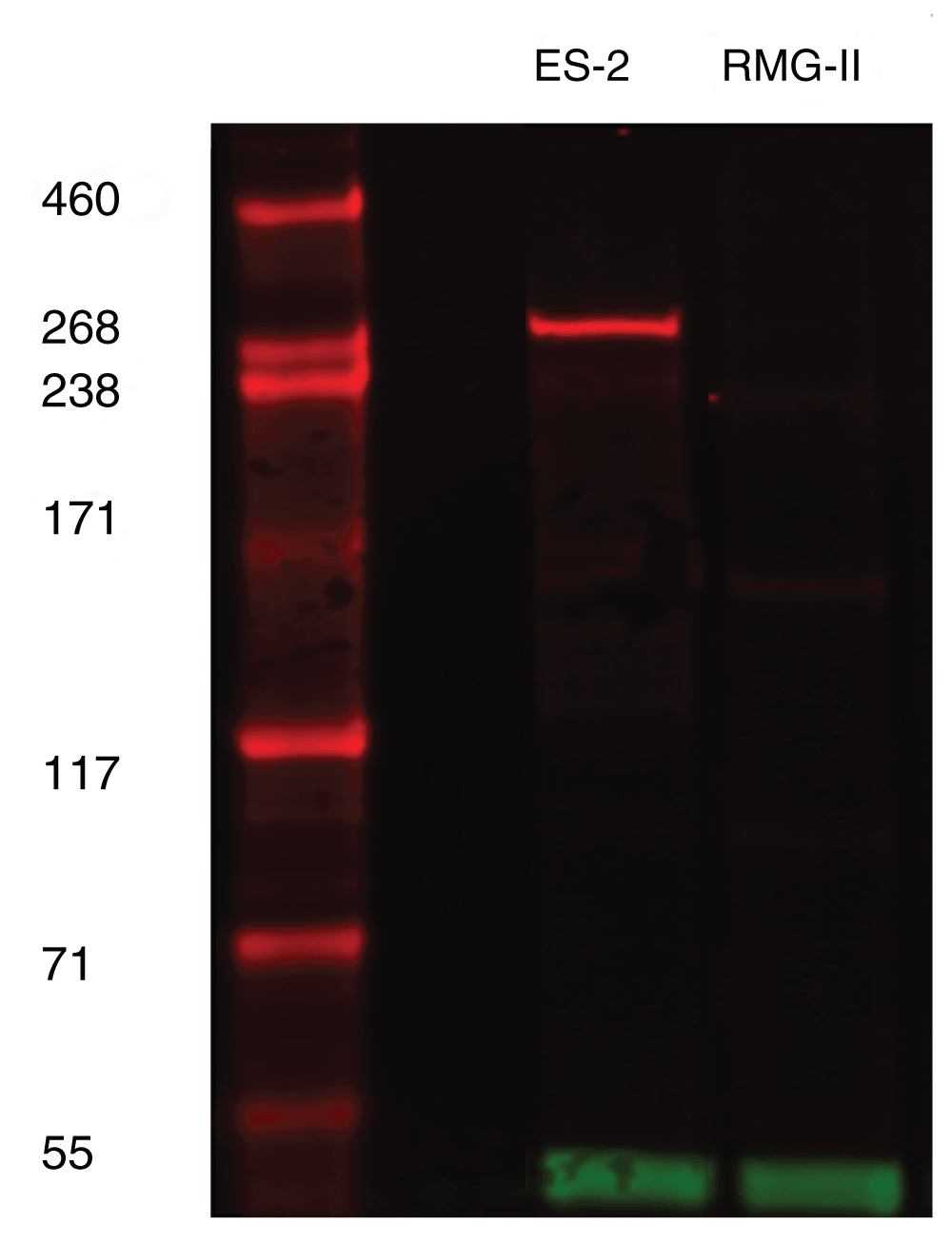
ARID1A (red band) can be seen to be present at approximately 270kD in ES2 cell line only. GAPDH at 37kD (green band) represents loading control.
For immunohistochemical validation, ES-2 and RMG-II cell lines were stained using three antigen retrieval conditions; ER1 (Sodium Citrate, pH6), ER2 (Tris/EDTA, pH9) and Enzyme 1 (Proteinase K, 100 µg/ml) at a fixed antibody concentration of 1 µg/ml. The enzyme retrieval demonstrated no nuclear signal for either ES2 or RMG-II cell pellets and was discarded for future work (Figure 2; Dataset a). The ER2 condition did demonstrate significant nuclear staining in the ES2 cell pellet with minimal background staining in the RMG-II cell pellet (Figure 2; Dataset b). However, the staining in the ER1 condition was determined to give the best signal:noise ratio with no background cytoplasmic staining and crisp nuclear staining for the cell pellet (Figure 2; Dataset c). Control slides, omitting the primary antibody, were negative except for the ER2 condition in the RMG-II cell pellet where a weak cytoplasmic background could be seen (Figure 2; Dataset d). Thus there was minimal background inherent in the staining procedure. It was therefore determined that the antibody showed specificity for formalin-fixed paraffin embedded tissues and could be run on murine tissue.

NPA denotes No Primary antibody control and represents the ER2 condition. Bar = 200 µm.
Murine uterine tissue was used as positive control tissue samples, given the literature data on cell lines and endometrial tissue. The ER1 condition at 1 µg/ml demonstrated clean nuclear staining in the uterine epithelial compartment as well as nuclear staining of stromal cells. However, the nuclear staining in the stroma was not universal and distinct negative nuclei could be seen (Figure 3; Dataset e). There was no cytoplasmic or extracellular stromal background staining present and the antibody titrated successfully losing the intensity of staining, as expected (Dataset e). Following this, a concentration of 0.5 µg/ml was used for future preparations which provided clear and consistent staining in repeated batches using a different antibody lot (Dataset f).
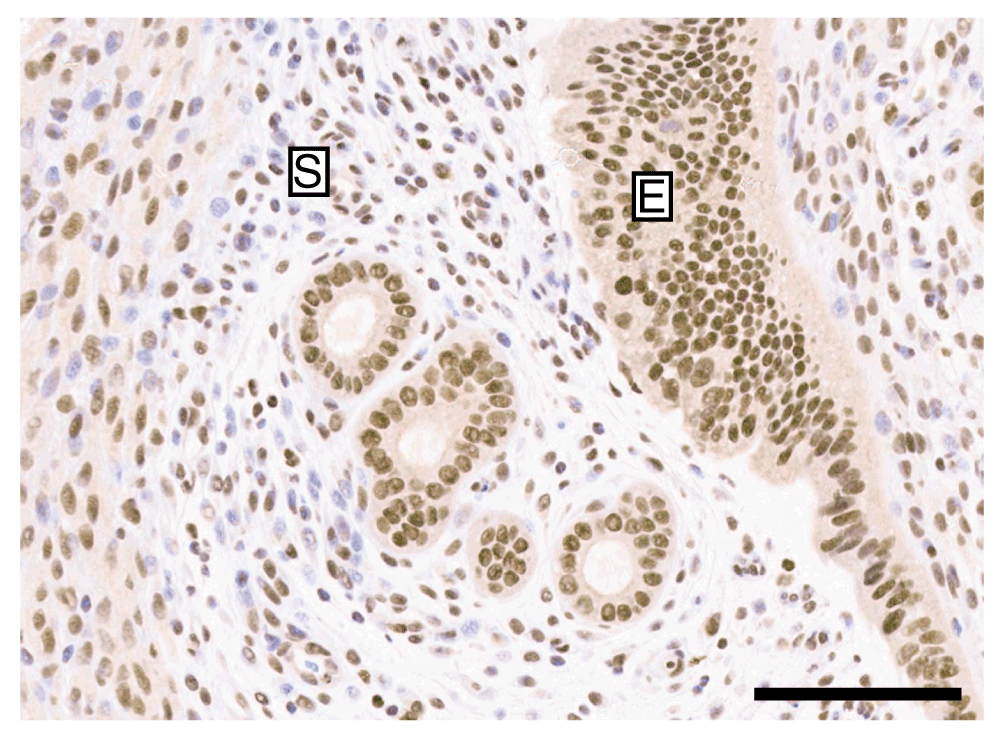
Murine uterine tissue stained by immunohistochemistry with anti-ARID1A using the ER1 condition and a concentration of 1 µg/ml demonstrating clear nuclear staining of the epithelial compartment (E) and negative nuclei in the stromal compartment (S). Bar = 100 µm.
Finally, when applied to a genetically engineered, tamoxifen-induced, Arid1a knockout mouse model, the staining in the uterine epithelium could be completely abrogated (Arrow, Figure 4b) when compared to the same area in a wild-type animal (Arrow, Figure 4a), while not affecting the staining in the stromal compartment.
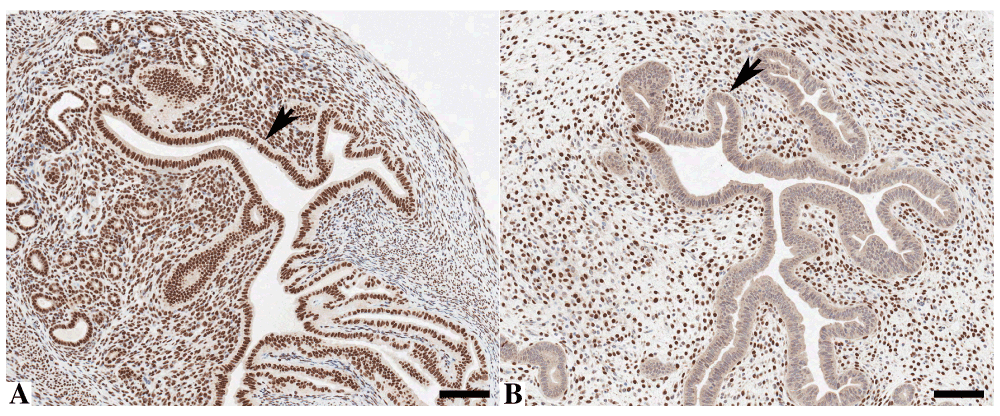
Murine uterine tissue stained by immunohistochemistry with anti-ARID1A demonstrating nuclear staining in wild-type mice (A) but loss of epithelial staining after ARID1A knock-out in Arid1afl/fl mice (B). Bar = 100 μm.
It is clear from the use of ES2 and RMG-II cell lines that the Atlas Antibodies ARID1A antibody is specific for ARID1A in both Western blots and formalin-fixed paraffin embedded preparations of human origin and, coupled with the literature evidence, that it is validated in human tissue.
The staining pattern when applied to murine uterus showing a clear nuclear pattern, is again consistent with the literature on this protein. Crucially, however, when stained on an ARID1a knockout mouse model, the staining could be almost completely abrogated in the epithelial compartment and thus when taken in combination, it is clear that the anti-human ARID1a antibody is cross-reactive with murine tissue and can be used for this purpose.
F1000Research: Dataset 1. Whole slide images from antibody validation of HPA005456 for immunohistochemistry, 10.5256/f1000research.5514.d3676110
WH wrote and conceived of the article, IG performed the Western blots and mouse experiments and requested validation of ARID1A in murine tissue, JM performed all immunohistochemical staining.
WH and JM are funded by Cancer Research UK, IG is funded by an MRC Clinical Research Training Fellowship, grant number G1001957.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would like to acknowledge the University of Cambridge, Cancer Research UK and Hutchison Whampoa Ltd. The authors would also like to acknowledge the staff at the Histopathology/ISH core facility at the Cancer Research UK Cambridge Institute for their assistance in preparing materials for this publication.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 3 (revision) 04 Jun 15 |
||
|
Version 2 (revision) 06 Jan 15 |
read | read |
|
Version 1 15 Oct 14 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)