Keywords
DNA
This article is included in the Bioinformatics gateway.
This article is included in the BioJS collection.
DNA
General compositional features of DNA sequences (e.g., GC/AT content) are important to understand the evolution, structure and function of genomes1. Genes are often characterised by having a higher GC content than the rest of the genome. It has also been shown that the length of the coding sequence is directly proportional to a higher GC content2. Stop codons, however, have a bias towards A and T nucleotides and are thus indicative of shorter gene sequences3.
GC content is usually expressed as a percentage value and sometimes as a ratio (GC ratio). The AT and GC content percentages and ratios can be measured in different ways (e.g., simple arithmetic or using a free online AT/GC calculator4).
To calculate the GC content percentage the following formula can be used5:
The AT content percentage can be calculated in a similar manner:
DNAContentViewer has been developed as a part of the BioJS project6. It is available via GitHub or the BioJS registry7, a centralised repository of BioJS components hosted at the European Bioinformatics Institute (EBI).
DNAContentViewer uses the JavaScript library D3.js8 for generating graphs. It reads the DNA sequence in raw format and plots it as a Scalable Vector Graphics (SVG) object. To our knowledge, this is the first client-side modular BioJS component that visualises GC/AT content directly from a raw sequence.
The way DNAContentViewer works is by calculating GC/AT content for dynamically generated slices of the sequence. This dynamic feature makes it fully scalable, working with sequences of any length. DNAContentViewer represents AT and GC content in separate graphs, allowing toggling of both to show and hide.
DNAContentViewer shows a percentage marker, which can be set on the left (by default), in the middle or on the right side of the graph. To adjust the visual aspect of the DNA content, DNAContentViewer contains simple controls for zooming and panning. DNAContentViewer can interact with other components via events and methods. Both events and methods are demonstrated through the component’s page in the BioJS registry. Following the BioJS standard recommendations, documentation is available giving more details on how to implement the extension of the component via the registry.
The code below shows how to initialize DNAContentViewer with some minimal input. The name given to YourOwnDivId constitutes the component container identifier and file is defined absolute or relative path to the file containing the raw DNA sequence.
var instance = new Biojs.DNAContentViewer({
target: "YourOwnDivId";
dataSet: "file"
});An example is shown in Figure 1, depicting GC/AT content from a genomic sequence in scaffold 1 of an assembly of the Chinese Hamster Ovary (CHO) genome. The GC/AT density data calculated from the raw sequence file are presented as bins, where the Y-axis indicates the percentage of GC/AT content for each bin. The overall AT content is higher than the GC content. The 13K region shows a much higher AT content than average. This may be indicative of a stop codon and a poly A tail present in the region. The figure also shows a dramatic change in the GC/AT content in the 23K-32K and 284K regions, showing that in these locations GC/AT content does not sum up to 100% of the total base content. This is indicative of regions that contain unidentified nucleotides (X or N bases) instead of A, T, G or C.
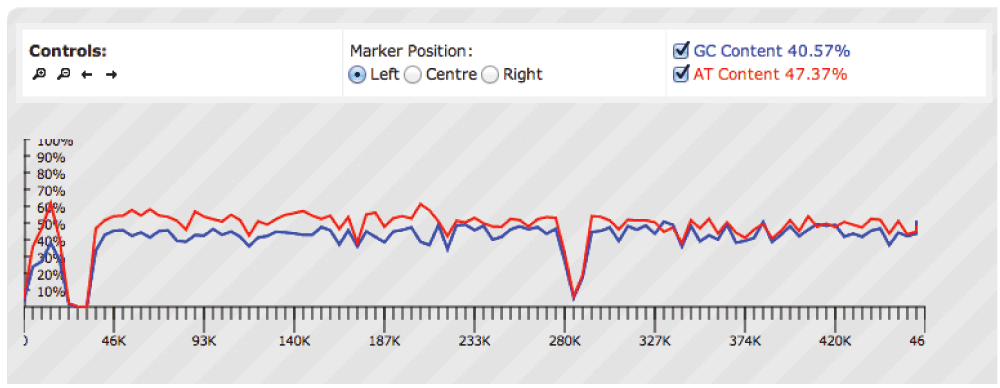
The top controls are designed to scroll sideways and to zoom; radio buttons can be used to set the position of the marker to the left (default), centre or to the right. Checkboxes can be used to toggle the GC/AT graph. Peaks show the GC/AT percentage density of dynamically generated bins. Differences can be observed around the 13K mark, with greater density of AT content, indicative of a stop codon and a poly A tail.
The DNAContentViewer component provides the functionality to visualise GC/AT content of DNA sequences within the BioJS framework. DNAContentViewer can be easily integrated with other BioJS web components or extended to provide new functionality. Like any other BioJS component, DNAContentViewer requires little technical knowledge for its reutilisation.
Zenodo: DNAContentViewer: a BioJS component to visualise DNA Content, doi: 10.5281/zenodo.77229.
GitHub: BioJS, http://github.com/biojs/biojs.
AT developed the code for DNAContentViewer and AT, MC and RD wrote the paper. SC designed the algorithm and wrote the initial version.
AT, SC, MC and RD were supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) National Capability Grant (BB/J010375/1) at TGAC.
We are grateful to all BioJS developers who have contributed their work under an open source license.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 13 Feb 14 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)