Keywords
Sry RNA, miR-138, circRNA, ceRNA, sponge activity
Sry RNA, miR-138, circRNA, ceRNA, sponge activity
Crosstalk involving RNA–RNA interactions adds a new dimension to our understanding of complex regulatory networks and offers profound implications for the elucidation of gene function1.
MicroRNAs (miRNAs) are a type of endogenously expressed small regulatory non-protein-coding RNAs that negatively regulate gene expression by base-pairing (with imperfect complementarity) to miRNA response elements (MREs), which are usually located within the 3′-untranslated region (3′-UTR) of target RNA transcripts2. According to their number and location, it has become evident that a biological process may involve multiple miRNAs, and that a given gene may be regulated by more than one miRNA. Salmena et al. coined the term “competitive endogenous RNAs” (ceRNAs) to designate those transcripts that may cross-regulate each other by competing for shared miRNAs3. Multiple classes of non-coding RNAs (long ncRNAs) including circular RNA (circRNAs) and pseudogenes, and protein-coding mRNAs function as key ceRNAs and “super-sponges” to regulate the expression of mRNAs in plants and mammalian cells4.
Recently, Hansen et al. and Memczak et al. described a new class of post-transcriptional regulatory RNAs that behave as circular endogenous RNA sponges (circRNAs) in two back-to-back papers published in Nature5,6. In both reports, the authors demonstrated that a ~1.5-kb single-stranded antisense circRNA molecule (human CDR1as or ciRS-7) containing multiple miR-7 binding sites densely arranged, acts as a natural miRNA sponge, by capturing complexes formed by miR-7/Ago2. Memczak et al. observed that human CDR1as expression in zebrafish impaired midbrain development, similar to knocking down miR-75.
Hansen et al. also showed that another circular RNA molecule, transcribed from the mouse Sry gene, could also act as an endogenous sponge. They noted that this transcript contains 16 binding sites for miR-138 and demonstrated in vitro that the Sry circRNA selectively “absorbs” this specific miRNA. Recently, Kartha and Subramanian asserted, based on the report by Hansen et al., that this Sry RNA is an antisense circular transcript that functions as a miRNAs sponge7. Although this apparently is a typographical error (antisense instead of sense), it was also referred as such in the original report by Memczak et al. in Nature. This suggests that the circular Sry transcript is, as occurs with the CDR1as sponge, an antisense circular RNA. Although it seems obvious that sponges are antisense to the miRNA they bind to, it should not be assumed that all circRNAs are transcripts in an antisense orientation to a protein coding gene, as occurs with CDR1as. A special feature of the Sry gene is that it can generate linear as well as circular transcripts depending on the use of alternative promoters (proximal vs distal)8. Capel et al. reported for the first time that the circular Sry RNA is derived from a sense sequence that consists of a single exon. This molecule is formed by the processing of a longer precursor transcript that contains one inverted repeat at each end. This unusual configuration promotes the formation of a stem-loop structure that facilitates the nucleophilic attack of a donor splicing site at the 3′ end to an acceptor site at the 5′ end, which results in its circularization8 (Figure 1). Thus, it can be asserted that this is, in fact, a circular sense Sry mRNA. Although the notion that the Sry circRNA is derived from an antisense transcript does not alter the interpretation of the results obtained by Hansen et al., we consider that this distinction is important, because it implies that both sense (e.g. Sry RNA) and antisense (e.g. CDR1as) transcripts could be circularized and act as RNA sponges, an observation which is not acknowledged by the authors of either of the original papers. Nevertheless, if the circular version of the Sry transcript can soak up miRNAs, can the Sry linear transcripts also do the same?
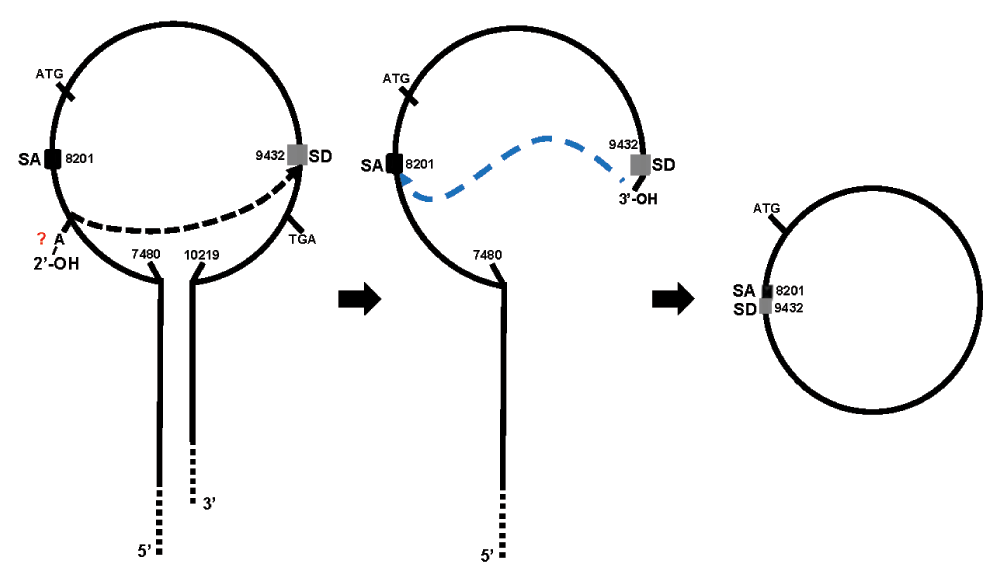
After the Sry pre-RNA is transcribed, a stem-loop structure is created due to the presence of inverted repeats at the 5′ and 3′ ends. A normal splicing reaction takes place when the splice donor (SD) is attacked by a 2′-OH, presumably from a branch site adenosine residue (A) located in the intron, causing the first cleavage of the phosphodiester backbone. The newly formed 3′-OH at the SD, attacks the 5′-P at the splice acceptor (SA) site, resulting in excision of the intron and ligation of the circular exon of 1231 nucleotides. Modified from Capel et al.8.
In this respect, there is evidence that certain miRNAs may function by targeting sites in the 5′-UTR9 and open reading frame (ORF) regions of mRNAs10, suggesting that miRNAs may modulate gene expression by mechanisms different from canonical 3′-UTR target mRNA suppression. Binding of a miRNA to a ceRNA not only prevents that miRNA from binding to other MREs, but can also repress translation from the coding segment of the ceRNA11. A study of the pseudogene of the phosphatase and tensin homolog PTEN, PTENP1, provided the first experimental evidence for the cross-talk between coding and non-coding RNAs12. Tay et al. found that several endogenous protein-coding transcripts, such as serine incorporator 1 (SERINC1), vesicle-associated membrane protein associated protein A (VAPA), CCR4-NOT transcription complex and subunit 6-like (CNOT6L), act as PTEN ceRNAs, which regulate PTEN tumor suppressor levels in a miRNA-dependent manner12. This clearly suggests that mRNAs can function as ceRNAs and we propose that the mouse linear Sry sense transcript could also behave as a miRNA sponge, or as a ceRNA for miR-138. The extent to which other animal or human antisense or sense circRNAs also behave as miRNA sponges will doubtlessly be a subject of intense research. Shortly after the emergence of circRNAs, the first public circRNA database (circBase version 0.1) was developed by the Rajewsky laboratory as a compendium of thousands of circRNAs sequences that are expressed in eukaryotic cells. Access to this resource allows us to use the information in order to validate those circRNAs that are probably involved in many important cellular processes. Nevertheless, the precise molecular mechanisms that underlie post-transcriptional repression by circRNAs remain still largely unknown, but their discovery demonstrates the importance of this distinct type of non-protein-coding regulatory RNAs for the elucidation of gene function. Moreover, due to their longer half lives in vivo, circRNAs may possess a great potential for therapeutic intervention. Thus, manipulating miRNA function, either by mimicking or inhibiting ceRNAs implicated in several disorders such as cancer, could provide a novel strategy to interfere with disease initiation and/or progression. The antisense modulation of circRNAs/ceRNA→miRNAs→mRNAs→protein regulatory networks could offer ingenious decoy combinations (antisense technology) as well as delivery platforms for concurrently target multiple miRNAs in abnormal or undesired conditions13.
JTGR and GAJ contributed extensively to this work and were involved in the critical revision of the manuscript. Both authors have agreed to the final version of the manuscript.
Work in the group’s lab is supported by grants CB-168661 from the Mexican Council of Sciences and Technology (CONACyT) and Mexican Federal Funds (HIM/2012/010-SSA 1017).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 13 Nov 14 |
read | read |
|
Version 1 11 Apr 14 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)