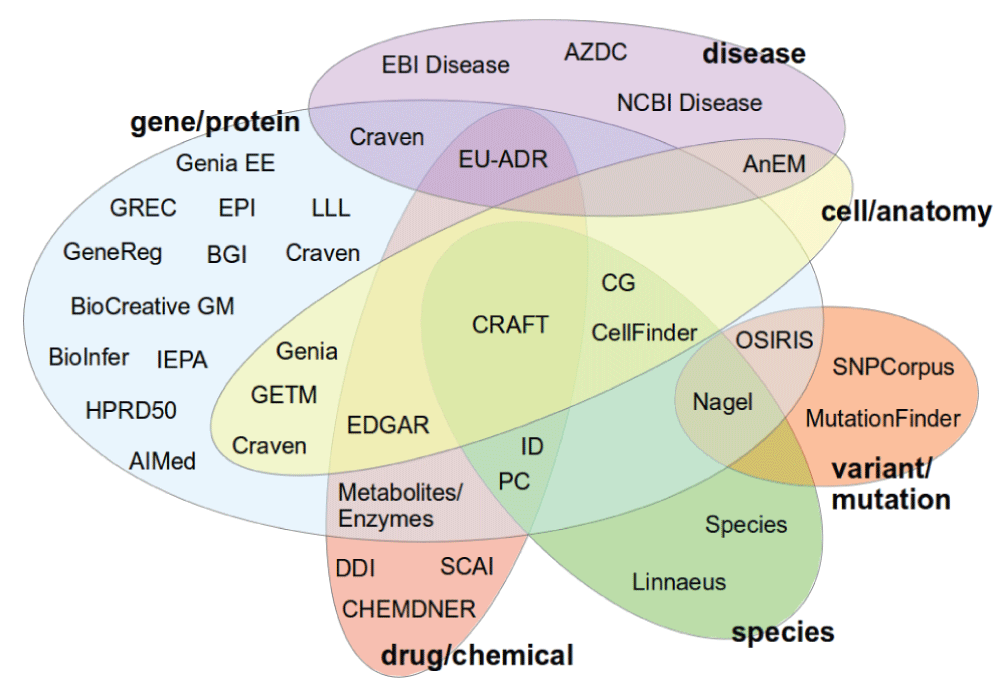
List of biological corpora
Here I present the list of 36 corpora which have been considered in this study. For each of them, I include a brief description of its origin, which may include the type of documents it contains (abstracts and full texts), its annotation schema, tools which have been based on it, further extensions it has received and the number of citations its publications have received. Table 1 shows a summary of these corpora, including their first publications, year of release (according to the main publication), number of citations according to Google Scholar (as December-2013) and the corresponding URL. Some of the corpora I present here are included in the WBI repository (http://corpora.informatik.hu-berlin.de), which provides their full visualization using the Stav/Brat annotation tool8. The collections are presented in the alphabetical order.
Table 1. Overview of the corpora: main publication, year of publication, citations in Google Scholar (as December-2013) and the URL are shown for each corpus.
AIMed. The AIMed corpus21 contains annotation on proteins and protein-protein interactions (PPI) for 200 abstracts, which were selected from the documents for which curated annotations were found in the Database of Interacting Proteins (http://dip.doe-mbi.ucla.edu/dip/Main.cgi). The corpus is one of the five corpora widely used for the development of PPI extraction methods50 and thus, has been used for the development of a variety of PPI tools51.
AnEM. The recently published AnEM corpus22 contains a total of 500 documents which contains annotations on the following anatomical entity types: organism subdivision, anatomical system, organ, multi-tissue structure, tissue, cell, developing anatomical structure, cellular component, organism substance, immaterial anatomical entity and pathological formation. It is probably the largest manually annotated corpus on anatomical entities and has been used for the development of the AnatomyTagger tool52.
AZDC. The AZDC corpus23 contains almost 800 abstracts which includes the ones available in the EBI disease corpus (cf. below) and some from the Craven corpus (cf. also below). It contains annotations for diseases and normalization to UMLS unique concepts for some semantic subtypes and was used for the development of named-entity recognition tools for disease names, such as the recent DNorm system53.
Bacteria Gene Interaction. The Bacteria Gene Interaction (BGI) corpus24 was developed in the scope of the BioNLP Event Extraction Shared Tasks 2011 for assessing the extraction of genetic processes in Bacillus subtilis. It is derived from the LLL corpus (cf. below) for PPIs. This corpus has been extended for the Gene Regulation Network (GRN) task54 in the 2013 edition of the same challenge.
BioCreative 2 Gene Mention. The BioCreative 2 Gene Mention4 corpus has been used in two editions of the BioCreative challenges (http://www.biocreative.org/) to foster improvements for gene/protein extraction. It is composed of sentences, opposed to documents, which were derived from Medline documents and contains annotation on gene and protein, though it does not make distinction between them. Given that it has been used in one of the most popular challenges in the BioNLP community, several studies have used this corpus for the development of gene/protein extraction systems, such as BANNER55.
BioInfer. BioInfer25 is also one of the five popular corpora available for PPI50. It contains sentences derived from more than 800 documents and annotations are available for genes, DNA families or groups, proteins, protein complexes and protein families and groups. Just as the other five PPI corpora, the BioInfer corpus has been used for training and evaluation of several tools51.
CellFinder. The CellFinder corpus2 was developed in the scope of the CellFinder database (http://cellfinder.de/) and includes annotations for six entity types (anatomical parts, cell lines, cell types, species and cell components) for 10 full text documents in the stem cell research field. This corpus has been mainly used for the evaluation of named-entity recognition approaches for the above entity types in Neves et al.2,56.
Cancer Genetics. The Cancer Genetics (CG) corpus26 was constructed for the Cancer Genetics task in the BioNLP Event Extraction Shared Task in 2013 and includes annotations on the development and progress of cancer. The corpus is composed of 600 abstracts split into three datasets and events are composed of anatomical and molecular entities, as well as annotations for organisms.
CHEMDNER. The CHEMDNER corpus7 has been recently created in the scope of the CHEMDNER task in BioCreative IV for assessing performance of named-entity recognition tools for chemical compounds. It contains 10,000 abstracts split into training, development and test datasets and annotations for chemicals are classified in eight categories, such as systematic, formula or abbreviation.
CRAFT. The CRAFT corpus5 is a recent and very comprehensive collection of 97 full text documents which has been annotated with concepts, such as gene/proteins, species, cells and chemicals, from nine ontologies and terminologies. The authors have reserved 30 of the full texts for a text mining challenge that is going to be carried out in the near future.
Craven. The so-called Craven corpus27 is in fact a collection of three corpora which contains annotations on sub-cellular locations, PPIs and gene-disease associations. These corpora have been used for the development of methods for extracting the above binary relationships and support construction of knowledge bases.
Drug-Drug Interaction. The Drug-Drug Interaction (DDI) corpus28 includes more than 700 documents derived from Medline and DrugBank, and includes annotations for drugs and binary relationships between them. It has been already evaluated on two shared tasks11,57 and thus, has been extensively used for both training and evaluation for NER and relatiosnhip extarction tasks.
EBI Disease. The EBI Disease corpus29 is composed of 600 sentences selected from the Craven corpus (cf. above) which have been extended with associations to unique concepts in the UMLS terminologies.
EDGAR. The EDGAR corpus30 contains annotations for genes, drugs and cells, including binary relationships between genes and drugs, genes and cells, and drugs and cells.
Epigenetics and Post-translational Modifications. The Epigenetics and Post-translational Modifications (EPI) corpus31 was developed for the BioNLP Event Extraction Shared Task 2011 and contains 1,200 abstracts annotated with events related to epigenetic changes. Just like the Genia Event Extraction corpus (cf. below), it contains annotations for genes/proteins and annotations identified as “Entity” which might refer to a variety of entity types, such as cell locations or small molecules.
EU-ADR. The EU-ADR corpus32 was constructed in the scope of the EU-ADR project, which aimed to automatically process health records. The corpus contains a total of 300 abstracts which are split into three groups, each containing annotations for two entity types and binary relationships: drug-target, drug-disease and target-disease.
GeneReg. The GeneReg corpus33 is composed of 314 abstracts related to Escherichia coli and contains annotations of events for gene expression regulation. It has been created in order to allow its interoperability with the Genia corpus (cf. below) and other lexical resources, such as WordNet and the Specialist lexicon.
Genia. The Genia corpus3 is probably one of the most popular corpora in the biomedical domain and has been used for the development of many named-entity tools, such as ABNER58, and also to assess systems in a shared task59. It contains 2,000 Medline abstracts with annotations based on the Genia ontology for DNA, RNA, proteins, lipids, cells, tissues, body parts and cell lines, among others.
Genia Event Extraction corpora. The Genia Event Extraction (Genia EE) corpus34 has started from the annotation of 1,000 abstracts, half of the Genia corpus (cf. above), and was annotated with genes/proteins and biological events, such as gene expression and gene regulations. This version of the corpus was used for the BioNLP Event Extraction Shared Task which took place in 200960 and then extended with 15 full texts for the following edition of the challenge that took place in 201161. A new corpus composed of 34 full texts was constructed for the third edition of the shared task that took place in 201362. The corpora have been used for the development and comparison of a variety of systems for extracting events.
GETM. The GETM corpus35 is composed of 150 abstracts derived from the development dataset of the Genia Event Extraction corpus (cf. above). Relationships were annotated between the gene expression events and the annotations for cells and anatomical locations which were present in the original corpus. It was used for the evaluation of a rule-based relationship extraction system on gene expression events in cell locations.
GREC. The GREC corpus36 contains annotations for 240 Medline abstracts for events on gene regulation and expression related to ontologies, such as Gene Ontology and Sequence Ontology.
HPRD50. The HPRD50 corpus37 has been created in the scope of the RelEx system and contains 50 abstracts and annotations for PPIs. The corpus is also one of the five PPI corpora50 and has been used for the development of a variety of PPI tools51.
ID. The ID corpus31 was developed for the BioNLP Event Extraction Shared Task 2011 and contains 30 full text documents annotated with biomolecular mechanisms of infectious diseases. The corpus is split into three datasets (training, development and testing) and events are related to annotations of proteins, chemicals and organisms.
IEPA. The IEPA corpus38 is composed of more than 200 sentences extracted from Medline abstracts and is annotated with binary relationships between proteins. It is also one of the five popular corpora available for PPI50.
Linnaeus. The Linnaeus corpus6 contains 100 full text documents annotated with annotations for organisms, all linked to identifiers in NCBI taxonomy (http://www.ncbi.nlm.nih.gov/taxonomy). It was built for the development of the Linnaeus system, one of the state-of-art tools for the annotation of organism names.
LLL. The LLL corpus39 for PPI in Bacillus subtilis was release in the scope of the Learning Language in Logic (LLL) shared task and was later also included in the package of the five popular corpora available for PPIs50. The proteins are identified as agent or target in the relationships.
Metabolites and Enzymes. The Metabolites and Enzymes corpus40 contains annotations for metabolites and enzymes names in almost 300 abstracts and was used for the evaluation of dictionary-based approaches for the recognition of these entity types.
MutationFinder. The MutationFinder corpus41 is composed of 508 Medline abstracts annotated with mutations and it was used for the evaluation of the homonymous tool based on regular expression techniques.
Nagel. The Nagel corpus42 contains annotations for protein residues, species and mutations in 100 Medline abstracts which have been used for the evaluation of a system developed for the extraction of these triplets.
NCBI Disease. The NCBI Disease corpus43 is composed of almost 800 abstracts derived from the AZDC corpus (cf. above) split into three datasets for training, development and blind testing. Annotations are classified into categories, such as modifier and specific disease, and it has been used for the development of the DNorm tool53.
OSIRIS. The OSIRIS corpus44 contains abstracts annotated with genes and sequence variants and was used for the evaluation of a dictionary-based system developed for the extraction of the later. Annotations for genes are normalized to identifiers from the NCBI EntrezGene database (http://www.ncbi.nlm.nih.gov/gene).
Pathway Curation. The Pathway Curation (PC) corpus45 was created for the homonymous task in the BioNLP Event Extraction Shared Task 2013 in which participants were required to extract biomolecular events to support curation of pathways. It includes a total of 525 abstracts annotated with events which contain chemicals, gene, proteins, complexes and cellular components as arguments.
PICAD. The PICAD corpus46 is another less popular PPI corpus composed of more that 1,000 sentences which were assembled in the scope of the development of a tool for this purpose.
SCAI. The SCAI corpus47 includes 100 abstracts with annotations for chemicals and training and test datasets for the recognition of IUPAC names. This has been one of the most popular corpora for chemical named-entity recognition and has been used for the development of many tools, such as ChemSpot63.
SNPCorpus. The SNPCorpus48 contains almost 300 abstracts and annotations for protein sequence and nucleotide sequence mutations and it has been used by the authors for extraction of these mentions from the text and their association to identifiers in biological databases.
Species. The Species corpus49 has been recently built as an alternative to Linnaeus (cf. above). Instead of using full text documents, it aimed at providing more variability on the species names by using eight groups of 100 abstracts on the following categories: bacteriology, botany, entomology, medicine, mycology, protistology, virology, and zoology.
Comments on this article Comments (0)