Keywords
Auxin-binding protein 1, ABP1, BELAYA SMERT, , BSM, embryo-lethality, allelism, short read DNA mapping
Auxin-binding protein 1, ABP1, BELAYA SMERT, , BSM, embryo-lethality, allelism, short read DNA mapping
The plant hormone auxin plays a central role in plant growth and development. Sensing and interpreting the fluctuating cellular auxin levels is crucial for mediating the corresponding physiological and developmental responses (Enders & Strader, 2015; Grunewald & Friml, 2010). Currently, two main auxin receptor/co-receptor systems are known and have been proposed to activate a range of cellular responses; among them the Auxin Binding Protein 1 (ABP1) has been considered as a prime candidate for the extracellular auxin receptor (Grones & Friml, 2015).
The first notion of ABP1 was based on the auxin-binding activity at the plant cell surface and in the endoplasmic reticulum in crude membrane preparations of etiolated coleoptiles (Hertel et al., 1972). This binding activity was characterized over the next decade by detailed biochemical studies (Batt et al., 1976; Ray et al., 1977). ABP1 was firstly purified from maize coleoptile cells (Lobler & Klambt, 1985) as a soluble 22-kDa large glycoprotein and later its crystal structure was elucidated (Woo et al., 2002).
The biological function and importance of ABP1 has been investigated extensively. Early studies have demonstrated that APB1 is involved in the rapid regulation of the membrane potential and ion fluxes at the plasma membrane and that it mediates the auxin-induced cell swelling, cell elongation, and cell division (Braun et al., 2008; Steffens et al., 2001; Tromas et al., 2009; Yamagami et al., 2004). In the presence of auxin, ABP1 activates the H+ pump ATPase that acidifies the extracellular space, presumably triggering cell wall loosening (Cosgrove, 2000).
With the advent of the genomic era and Arabidopsis thaliana as a model system, genetic tools have been adopted to facilitate the studies of the ABP1 developmental roles. Using a reverse genetic approach, two independent Arabidopsis thaliana knock-out alleles of ABP1 gene were identified, abp1-1 and abp1-1s (Chen et al., 2001; Tzafrir et al., 2004) and both were reported to be allelic and embryo-lethal, arresting the embryo development at the globular stage. This defined ABP1 as an essential protein required from early embryonic development with functions in cell division and elongation (Chen et al., 2001). However, embryo-lethal phenotype of abp1 knock-out mutants has hampered its further functional characterization. This was rectified by the generation of transgenic lines conditionally downregulating ABP1 levels by ethanol-inducible immuno-modulation or antisense approaches that, despite entirely different technologies used, show the same phenotypes confirming the role of ABP1 in cell division and cell expansion (Braun et al., 2008; David et al., 2007).
Both downregulation lines along with the weak abp1-5 point mutation allele showed defects in clathrin-mediated endocytosis of PIN auxin export proteins (Dhonukshe et al., 2007; Petrášek et al., 2006) and their inhibition by auxin (Paciorek et al., 2005). In contrast, the ABP1 gain-of-function mutation has an opposite effect, promoting PIN internalization in tobacco cultured cells and stable Arabidopsis lines (Robert et al., 2010). Opposite effects of ABP1 loss- and gain-of-function lines were observed also for auxin effects on arrangements of microtubules (Xu et al., 2014) and for controlling morphogenesis and shape of leaf epidermal pavement cells (Nagawa et al., 2012; Xu et al., 2010). Furthermore, the importance of auxin binding to ABP1 for gain-of-function phenotypes has been demonstrated by analysis of ABP1 variants with introduced mutations in the auxin binding pocket (Grones et al., 2015). Thus various types of loss- and gain-of-function strategies show an internally consistent picture of ABP1 signaling being involved in a range of physiological and cellular processes.
An important breakthrough came with the finding that the auxin-bound ABP1 docks on the extracellular domain of the Transmembrane Kinase 1 (TMK1) (Dai et al., 2013; Xu et al., 2014) which added the missing piece to the puzzle of how the auxin signal is transmitted from the cell surface to the cytosol and further confirmed involvement of the ABP1/TMK1 pathway in auxin-mediated development, particularly in pavement cell morphology (Grones & Friml, 2015).
Surprisingly enough, shortly after these studies had been published, Gao et al. (2015) described two independent, full loss-of-function abp1 mutants of A. thaliana (abp1-c1 and abp1-TD1) that show no apparent developmental defects under normal growth conditions. This directly contradicts the embryo-lethal phenotypes of the originally described abp1-1 and abp1-1s lines (Chen et al., 2001; www.seedgenes.org/SeedGeneProfile?geneSymbol=ABP+1) and also questioned the relevance of the aforementioned studies.
The explanation of these contradictory results is therefore of crucial importance to understand the biological role of ABP1. In this study we aim to clarify the discrepancy between the dramatic embryolethal phenotype of the originally described abp1 knock-out mutants and the newly identified loss-of-function alleles.
Arabidopsis thaliana mutants used in this study were: abp1-1 (Chen et al., 2001), abp1-1s (NASC accession N16148), abp1-c1, abp1-TD1 (Gao et al., 2015), bsm1-1 (Babiychuk et al., 1997). A. thaliana Col-0 wild type seeds were obtained from The Nottingham Arabidopsis Stock Centre (NASC, http://arabidopsis.info). The seeds were vernalized for 3 days in the dark at 4°C. Plants were grown under long-day conditions (16-h light/8-h dark cycles) at 22°C in soil (Potgrond P) : perlit 4 : 1 substrate and watered regularly with tap water.
For selection of abp1-1, abp1-1s and bsm1-1 heterozygous plants, seeds were surface-sterilized with chlorine vapor and plated on 1/2 MS agar medium (pH 5.9) containing 1% (w/v) sucrose and 25 µg/mL kanamycin according to Harrison et al. (2006).
Data from two publicly available short read libraries (NCBI Short Reads Archive accessions SRX759525 and SRX703650) from A. thaliana re-sequencing experiments were downloaded and mapped to the deposited reference genome TAIR10 (accessions NC_003070, NC_003071, NC_003074, NC_003075, NC_003076) using the Bowtie 2 plugin within the Geneious software, version 7.1.7 (http://www.geneious.com). For the first analyzed short read library (accession SRX759525), mapping parameters were set to a local alignment (“--very-fast-local”) with multi-mapping allowed (value k = 10). For the second analyzed short read library (accession SRX703650), mapping parameters were set to end-to-end alignment and the best-match mapping option (k = 1) was used. In both cases the seed length (L) was set to 22 and number of mismatches (N) to 1. To maximize the mapping coverage short reads were mapped single-end. For all the remaining parameters the default Bowtie 2 values were used. For both alignments, the median coverage at the ABP1 locus (bases 1 319 656 - 1 321 477 of NC_003075) was compared to the surrounding 20 Kbp region (Figure 1A and 1B) and to the whole chromosome 4 (Figure 1C and Dataset 2 for SRX759525; Figure 1D and Dataset 3 for SRX703650). Mapping coverage is defined as the number of reads mapped to a given base. Coverage data from the Geneious software were exported and visualized using Matlab (v. R2011b).
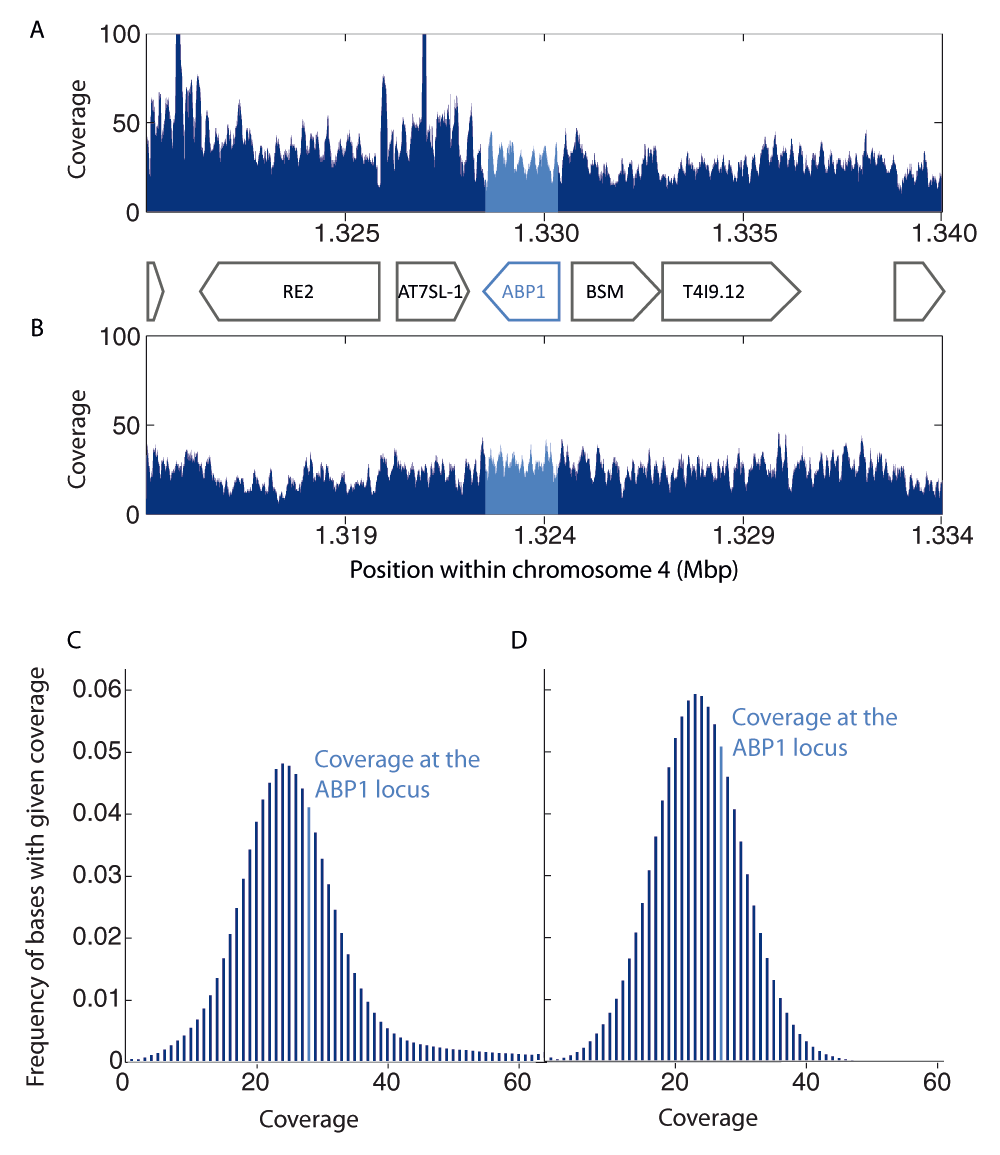
To test the hypothesis of hidden ABP1 duplicates in the genome of Arabidopsis thaliana, two short reads libraries from A. thaliana re-sequencing projects (NCBI accession numbers SRX759525 and SRX703650) were separately aligned to the reference Arabidopsis genome (TAIR10) using the BOWTIE 2 suit using different mapping parameter sets (see the Methods section). (A, B) The coverage of the 20 kbp consensus sequence within the chromosome 4 that surrounds ABP1 is shown for the two libraries: SRX759525 (A) and SRX703650 (B). (C, D) The overall distribution of the base coverage within the chromosome 4 is shown below. The median coverage at the ABP1 locus is highlighted in light blue and is well within the expected coverage values for both SRX759525 (C) and SRX703650 (D).
The T-DNA insertional mutants were genotyped by using a PCR-based method (Alonso et al., 2003). Amplification of PCR products was made using Phire Plant Direct PCR Kit (obtained from Thermo Scientific by Finnzymes, Espoo, Finland) following manufacturer’s instructions for the dilution protocol and using Bio-Rad T100 Thermal Cycler. The PCR conditions were as follows: initial denaturation for 5 min. at 98°C and subsequent 40 cycles: denaturation for 5 s at 98°C, annealing for 10 s at the respective annealing temperature for each primer set (calculated with the Thermo Scientific Tm calculator; available at www.thermofisher.com), elongation for 30 s at 72°C and final elongation for 1 min. at 72°C.
Genotyping primers were as follows: cdsBSM_F and nptII_R for the bsm mutation, ABP1_3UTR_FOR and WiscDsLoc_REV for the abp1-1 mutation, WiscDsLoc_REV and qBSM_R2 for the abp1-1s mutation. Genotyping of abp1-c1 and abp1-TD1 mutants was described previously (Gao et al., 2015). Sequences of all primers used in this study are listed in Table 1. Purified PCR products from plants positive for the presence of the T-DNA insertion were sequenced using a commercial service (LGC genomics, www.lgcgenomics.com). Primers used for sequencing were the same as for PCR. Sequence reads were aligned against A. thaliana genome (TAIR 10) using the BLAST tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The position of the mutation was identified at the border of the sequence part aligned to ABP1 locus and the sequence part aligned to the transformation vector. Position of individual mutations was mapped to the sequence of ABP1/BSM locus acquired from the TAIR database (https://www.arabidopsis.org), visualized with the SnapGene Viewer software version 2.8.2 (http://www.snapgene.com/products/snapgene_viewer) and edited in MS Power Point 2010.
For the RNA extraction approximately twenty 8 day-old seedlings were frozen in liquid nitrogen. Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and purified using RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions. For digestion of genomic DNA the following modification was incorporated into the protocol: in step 7, first 350 µL of RW1 buffer was added to the RNeasy spin column and centrifuged (8000 rpm, room temperature, 15 s). Subsequently, 40 µL of the RNase-free recombinant DNase I incubation mix (mixture of 5 µL DNase I and 35 µL buffer RDD) (Roche) was added to the column and incubated for 15 min at room temperature. Next, another 350 µL of RW1 buffer was added to the column and centrifuged again (8000 rpm, room temperature, 15 s). Two µg of purified DNase I pre-treated total RNA was used for a reverse transcription reaction using the iScript cDNA Synthesis Kit (BioRad, Hercules, CA, USA). Primers used for the quantitative RT-PCR were designed using QuantPrime (http://www.quantprime.de). For amplification of the BSM cDNA fragment (88 bp in length) primers qBSM_F2 and qBSM_R2 were used. The ABP1 cDNA fragment (84 bp in length) was amplified with A2E and ABP1-586R primers. Arabidopsis tubulin beta chain 2 (TUB2, At5g62690) was used as a reference gene and amplified with TUB-2_F3 and TUB-2_R3 primer set (Table 1, Dataset 1). All primers were synthesized by commercial service (https://www.eurofinsgenomics.eu/). qRT-PCR was performed using the LightCycler 480 SYBR Green I Master chemistry (Roche) in a LightCycler 480 II thermal cycler (Ser. no. 5659, Roche) according to manufacturer’s instructions. A 1:10 cDNA dilution was used as a template to prepare 5 µL reaction mixture (final volume). The cycling conditions were as follows: pre-incubation for 10 min. at 95°C and subsequent 45 cycles: denaturation for 10 s at 95°C, annealing for 15 s at 60°C, elongation for 15 s at 72°C followed by high resolution melting analysis. Gene expression was calculated with the 2-ΔΔCT method (Livak & Schmittgen, 2001). Individual experiments were made in a technical triplicate.
To compare the stage of embryo arrest, seeds from siliques of the heterozygous abp1-1 and bsm plants in the 7th to 8th developmental stage were used. For evaluation of embryo development, seeds from the siliques in the 3rd to 5th developmental stage were used. Siliques were dissected using a sharp needle and seeds were extracted and transferred onto microscopic slides into the drop of the Hoyer’s solution (http://www.seedgenes.org/Tutorial.html) and cleared for 1 h similarly as described (Friml et al., 2003). The embryos were analyzed under 20 × magnification using a digital camera system of the Olympus BX53 microscope by Normanski optics (Differential Interference Contrast (DIC) light microscopy). Images were processed in the ImageJ software, v. 1.48 (http://imagej.nih.gov/ij/) and mounted in Adobe Illustrator CS5.1.
Approximately 4 week-old plants were used for crossing. Flowers of recipient plants were emasculated 2 days before pollination. Crosses were generated by manual pollination. Green siliques were dissected 8 days after pollination and number of white and green seeds was counted and their ratio calculated.
One of the possibilities explaining at least some of the recent ABP1 controversies, in particular the strong phenotypes of the conditional lines versus no apparent phenotypes of the abp1-c1 and abp1-TD1 alleles, is the presence of a second, non-annotated ABP1 gene copy in the genome of A. thaliana that is functionally redundant and not disrupted in the abp1-c1 and abp1-TD1 alleles. Highly similar copies of the ABP1 gene are present in the annotated genomes of maize (Zea mays), rice (Oryza sativa) or poplar (Populus trichocarpa) (http://phytozome.jgi.doe.gov). During genome assembly, highly similar sequences such as transposons or recently duplicated genes could collapse into one copy resulting in no annotation of the duplicated copy (Stephane Rombauts, personal communication, 30th January 2015). A non-annotated second copy of the ABP1 gene is also present in the sequenced genome of the moss Physcomitrella patens (http://phytozome.jgi.doe.gov) raising the possibility that a similar situation could take a place in the A. thaliana genome.
We hypothesized that in case of a second non-annotated copy of ABP1 in the A. thaliana genome, the short reads coverage at the ABP1 locus would be doubled compared to the neighboring DNA regions and to the coverage of most other loci on the chromosome. To address this question we mapped short reads from two publicly available short read libraries from A. thaliana re-sequencing projects to the annotated Arabidopsis reference genome (TAIR 10) and analyzed the coverage at the ABP1 locus.
As depicted (Figure 1), the number of aligned short reads at the ABP1 locus is comparable with the short read coverage of the neighboring sequences and is well within the range of most common coverage values for loci on chromosome 4 indicating that no hidden copy of ABP1 is present in the A. thaliana genome.
Another possibility to explain phenotypic discrepancies between different abp1 mutant alleles is a disruption of another gene in addition to ABP1 that would be responsible for the strong phenotypes in the abp1-1 and abp1-1s alleles. As it can be seen on the ABP1 locus map (Figure 2A), the ABP1 gene is closely located to its neighboring gene BSM in a head-to head orientation with translational start codons and 5’-UTR regions of the two genes being just 708 bp and 381 bp respectively apart from each other. BSM encodes the mitochondrial transcription termination factor (mTERF)-like protein responsible for mitochondrial transcription termination in humans. In plants, mutants disrupting the BSM gene have been shown to be embryo-lethal (Babiychuk et al., 2011), therefore we suspected that the BSM gene is the most likely off-target of mutations in the abp1-1 and abp1-1s alleles.
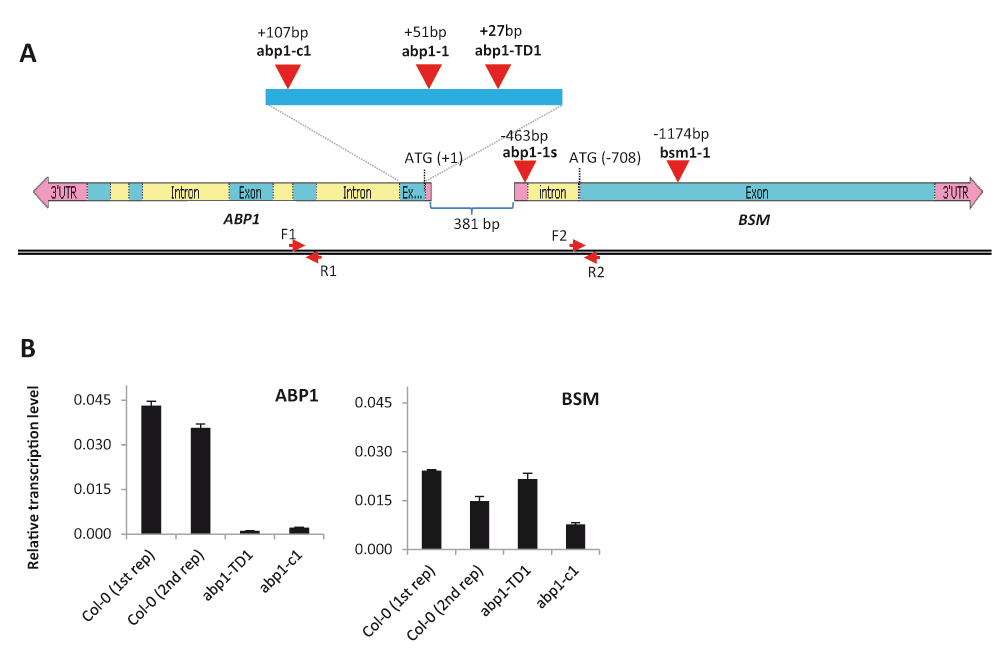
(A) Map of the ABP1/BSM locus showing the mapped position of existing loss-of-function mutant alleles (red arrowheads) relative to the ABP1 translation start. Note that the T-DNA insertion in abp1-1s allele is located in the 5’-UTR region of the BSM gene. Positions of the forward primers ABP1-586R (F1) and qBSM_F2 (F2) and reverse primers A2E (R1) and qBSM_R2 (R2) used for the qRT-PCR are indicated by red arrows. (B) Quantitative real-time PCR of the ABP1 (left graph) and BSM (right graph) shows relative transcript levels in wild-type Col-0, abp1-c1 and abp1-TD1 seedlings. The reference gene was TUB2 (At5g62690). The transcript levels were calculated using 2-ΔΔCT method. The data represent average relative quantity values of three technical replicates, and the bars denote standard errors. For wild-type Col-0 plants data from 2 biological replicates are shown.
To look into this possibility closely, we mapped the relative position of individual mutations within the ABP1/BSM locus (Figure 2A). Surprisingly, we found that the T-DNA insertion in abp1-1s mutant is in fact located in the 5’-UTR of the BSM gene. Because ABP1 and BSM share a common promoter region, we hypothesized that the T-DNA insertion in abp1-1 could also disrupt the expression of the BSM gene thus causing the described embryonic lethality. Notably, the T-DNA insertion in the wild type-like allele abp1-TD1 is located just 24 bp apart from the abp1-1 T-DNA insertion and is even closer to the BSM gene. We speculated that the dramatic phenotypic difference between both closely positioned T-DNA-based insertions might be caused by the presence of multiplied 35S enhancers within the T-DNA construct in the abp1-TD1, because this allele was recovered from the activation tagging Arabidopsis mutant collection (SASKATOON) and harbors multiplied enhancers near the right T-DNA border on the inserted T-DNA element (Figure 2B).
To see whether the expression of the BSM gene in homozygous abp1-TD1 plants was affected by the T-DNA insertion, we performed a qRT-PCR analysis. Surprisingly, the BSM transcript levels in abp1-TD1 were not substantially affected by the T-DNA insertion and were comparable to wild type Col-0 plants (Figure 2B). On the other hand, the expression of the ABP1 gene was abolished in both abp1-c1 and abp1-TD1 homozygous plants as reported previously (Gao et al., 2015). For abp1-1, abp1-1s or bsm mutants it was only possible to evaluate the BSM expression in the heterozygous plants (due to the lethality of homozygous mutants) and this showed no substantial differences compared to wild type plants (Dataset 1).
Therefore, it is possible that while in the abp1-TD1 allele the expression of the BSM gene is rescued by its ectopic expression driven by 35S enhancers within the T-DNA, in the abp1-1 allele, where no such a mechanism exists, the expression of both genes may be disrupted. In addition, the abp1-c1 null allele only contains a small 5 bp deletion at the end of the first exon of ABP1 which is less likely to have any effect on the BSM gene compared to a few kB long T-DNA insertions in other alleles. Thus it is plausible that in abp1-1 and abp1-1s, both ABP1 and BSM genes are disrupted.
To compare the embryo-lethal phenotypes of abp1-1 and abp1-1s alleles with the embryo-lethal phenotype of the bsm1-1 T-DNA insertional knock-out allele, we analyzed the embryo development in siliques of abp1-1, abp1-1s and bsm1-1 heterozygous plants along with homozygous new abp1 knock-out alleles.
The inactivation of BSM gene by T-DNA insertion has been shown to cause an embryo arrest at the late globular stage of development (Babiychuk et al., 2011). In the abp1-1 mutants the embryo arrest has been reported at the early globular 32-cell stage in 25% of immature seeds (Chen et al., 2001). In both cases the homozygous mutant embryos never reach the heart-shape stage, indicating prevention in vertical elongation and subsequent anticlinal placement of division planes in the lower tier of cells. Other characteristic features reported in abp1-1 embryos have been connected with failure in degeneration of the suspensor structure and ectopic anticlinal divisions of basal cells resulting in longer suspensors at the later stages (Chen et al., 2001).
Inspection of embryos at different stages from the homozygous abp1-c1 and abp1-TD1 plants did not reveal any aberrations in the embryo development as compared to the wild type embryos (Figure 3) consistent with the lack of obvious postembryonic phenotypes (Gao et al., 2015). Also, no white aborted seeds were found in the older siliques of these plants (Supplementary Figure 1).
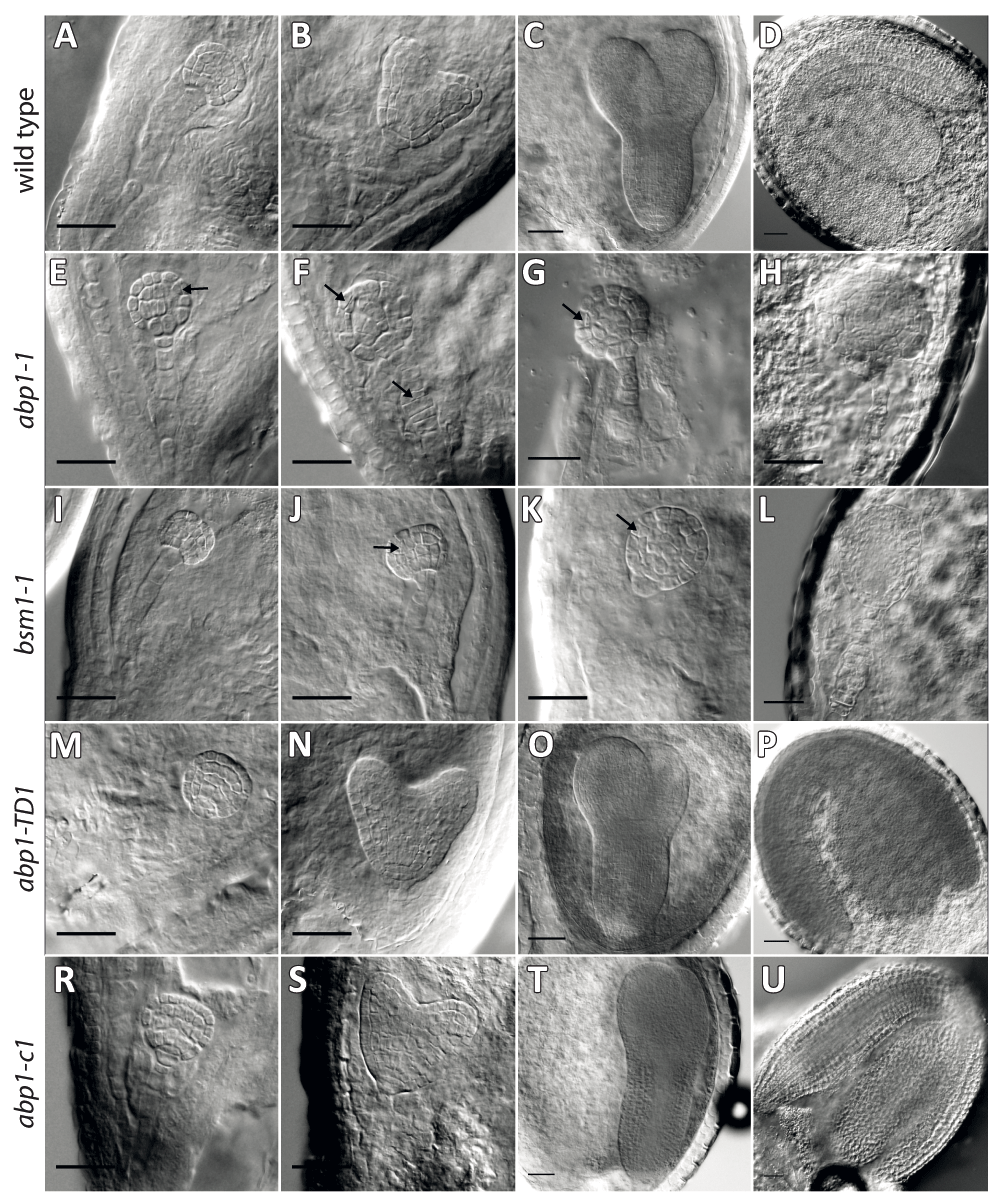
Seeds from the siliques of abp1-1+/- (A-H), bsm+/- (I–L), abp1-TD1 -/- (M–P), and abp1-c1 -/- (R–U) plants at different developmental stages were isolated and cleared in Hoyer’s solution and screened for defects in embryo development using Normanski optics. Wild type-looking embryos in the seeds of abp1-1 mutant (A–D) showed normal developmental progression: the late globular stage (A), early heart (B), torpedo (C) as well as the mature embryo stage (D). At the globular stage we started to observe differences in embryo development between different mutants earliest manifested by periclinal divisions in protodermal cells (arrows) (A, E, I, M). When most of the wild-type embryos entered the early heart stage (B) some abp1-1 embryos, from the same silique as (B) showed abnormal cell divisions (F), also visible in bsm (J) (arrows) but not in the abp1-TD1 (N) or abp1-c1 mutant (S). At the later stages of embryo development, when wild-type embryos in abp1-1 (C, D) as well as embryos in abp1-TD1 (O, P) and abp1-c1 (T, U) reached the torpedo (C, O, T) and mature stage (D, P, U), the mutant abp1-1 (G, H) and bsm (K, L) embryos were still arrested at the late globular stage. While the bsm mutant embryos show signs of apical basal polarity, abp1-1 embryos formed ball-shaped symmetric structure. Also, abnormal cell divisions in the suspensor cells were visible (arrows). Scale bars, 25 µm.
Opening and inspection of the older siliques in abp1-1, abp1-1s and bsm1-1 heterozygous plants did not reveal any visible differences between those alleles. In all three cases, the siliques carried approximately 25% of white aborted seeds (see Figure 4) containing developmentally arrested embryos (Figure 3). Analysis of the younger embryonic stages did not reveal any significant differences between the analyzed alleles. The obvious embryo defects at the early globular stage were infrequent, and in both abp1 and bsm mutants started to be more pronounced at the late globular stage manifested by the failure of directional cell elongation leading to ball-shaped embryos at later stages. Another characteristic feature for both mutants was a disrupted cell division pattern with frequent periclinal divisions of outer layer cells (Figure 3B). At the later stages of embryo development, bsm mutant embryos showed some degree of apical-basal polarity by forming sometimes oval-shaped structures while abp1-1 mutant embryos continue to divide non-directionally forming more symmetrically ball-shaped embryos. These differences could be explained either by different backgrounds of the two mutant alleles (C24 in the case of bsm and Wassilewskija in the case of abp1-1, respectively) or by simultaneous disruption of both ABP1 and BSM genes in the abp1-1 mutant that may produce stronger effect as compared to a single BSM loss-of-function mutation in bsm1-1. Despite these minor differences at the very late embryo stages, both mutations generated undistinguishable cell elongation and cell division pattern defects starting in both cases at the globular stage resulting in similar embryo-lethal phenotypes.
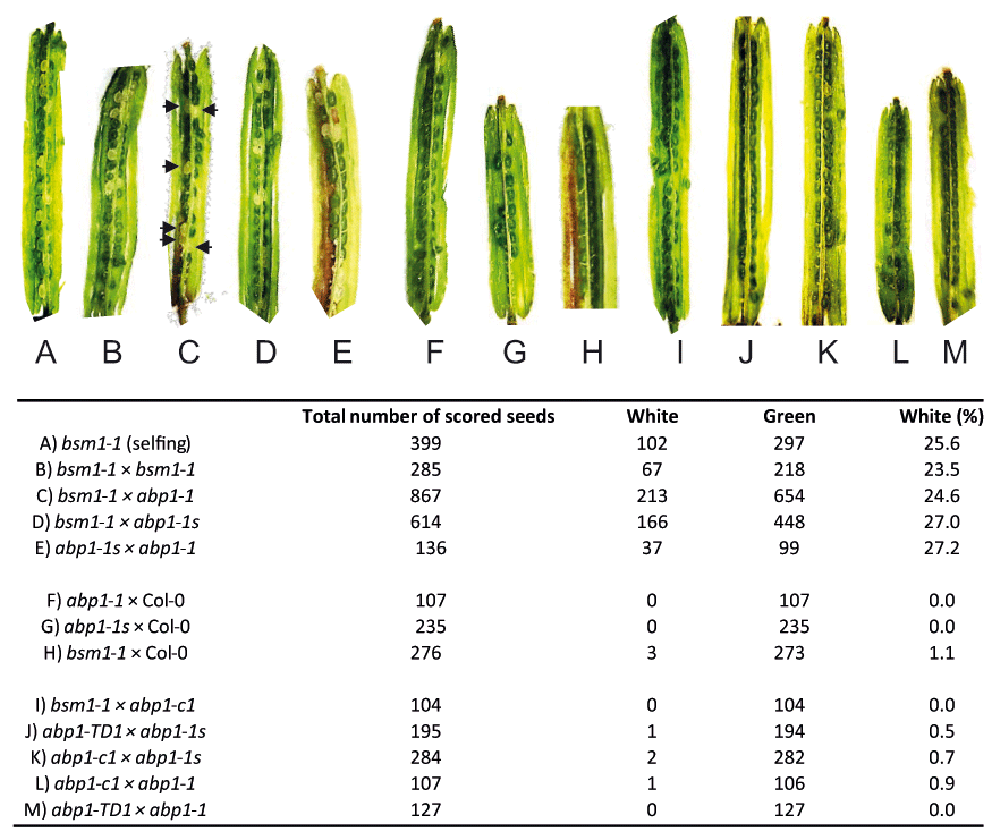
The embryolethal phenotype was detected in siliques 8 days after pollination by the presence of white seeds (arrested embryo development). It was not possible to complement the embryo-lethal phenotype of bsm mutant (A, B) with either abp1-1 or abp1-1s mutant (C, D). Similarly, abp1-1 and abp1-1s mutants did not mutually complement (E). As a control Col-0 plants were crossed into the abp1-1 (F), abp1-1s (G) or bsm (H) mutants showing complementation of the embryo-lethal phenotype. Genetic crosses of the recently identified abp1 null mutants (abp1-c1, abp1-TD1) with bsm (I), abp1-1s (J, K) or abp1-1 (L, M) lines resulted in complementation of the embryo-lethal phenotypes showing that in these lines the disruption of the BSM and not the ABP1 gene is responsible for the embryo-lethal phenotype. Arrows indicate the position of white seeds in (C). Number of scored seeds for each cross and the percentage of white seeds are listed in the table below. Note that in not complementing crosses, the segregation of embryo-lethal seeds follows the Mendelian genetic laws as expected.
As the embryo-lethal phenotypes of abp1 and bsm mutants are indistinguishable from each other, we tested whether these phenotypes might be due to the mutations affecting the same gene or they are independently embryolethal. To address this hypothesis we performed allelic crosses between the bsm and abp1-1 as well as abp1-1s line and looked for the presence of white seeds carrying defective embryos within immature siliques 8 days after pollination of emasculated flowers. We assumed that if the mutations affect the same gene it will not be possible to complement the embryolethal phenotype of abp1-1 or abp1-1s mutants with a functional ABP1 allele from the bsm mutant which will result in the segregation of approximately 25% not developing (white) seeds in the F1 generation.
After dissection of siliques of bsm1-1 × abp1-1, bsm1-1 × abp1-1s or control crosses bsm1-1 × bsm1-1 and abp1-1s × abp1-1, we observed the presence of clearly distinguishable white seeds (Figure 4A–E). The segregation ratio of 1:4 white:green seeds was observed (Table in Figure 4, Supplementary Table 1) which is in perfect agreement with the Mendelian genetic laws implying that abp1-1 and abp1-1s mutants do not carry a functional BSM gene. On the other hand, when crossed with wild type (Col-0), abp1-c1 or abp1-TD1 mutants, more than 99% of seeds in the siliques were green, indicating that a functional copy of the BSM gene present in these lines was able to complement the embryolethal phenotype of bsm1-1, abp1-1 and abp1-1s lines (Figure 4F–M).
In summary, these observations show that the described embryolethal phenotype of the abp1-1 and abp1-1s lines is caused by a disruption in the BSM gene.
Since its discovery, the biological importance of the ABP1 protein as a plasma membrane auxin receptor has been a matter of debate, in part because of its predominant ER localization in plant cells where the conditions for auxin binding are unfavorable (Habets & Offringa, 2015; Napier et al., 2002). Nonetheless, after the two independently isolated Arabidopsis abp1 loss-of-function alleles were reported to be embryolethal (Chen et al., 2001; www.seedgenes.org/SeedGeneProfile?geneSymbol=ABP+1), the crucial importance of this protein in cell division and elongation was accepted. This view was challenged by the isolation of two new knock-out alleles (Gao et al., 2015) that harbor mutations in a close vicinity to the previously published insertions and show no obvious phenotypes.
The phenotype of the originally reported abp1-1 mutant was reported to be complemented by the 35S::ABP1 overexpression construct (Chen et al., 2001). However, despite multiple attempts it was not possible to repeat this complementation either with constructs for overexpression of the wild type ABP1 copy or by the genomic fragment (Grones et al., 2015). Altogether, these findings implied that there exists another cause for the drastic phenotypes present in the abp1-1 and abp1-1s mutants.
Here we show that the neighboring gene BSM, which has been shown previously to be crucial for embryogenesis in Arabidopsis (Babiychuk et al., 2011) is likely to be also disrupted by the T-DNA insertions in the original abp1-1 and abp1-1s mutants, but not in the newly isolated loss-of-function lines abp1-c1 and abp1-TD1. Furthermore, we demonstrate that the mutant embryos of abp1-1 and abp1-1s as well as the loss-of-function bsm1-1 allele showed similar embryo phenotypes and are arrested at the globular stage as it was shown in the original studies (Babiychuk et al., 2011; Chen et al., 2001).
By allelic complementation experiments we showed that the embryolethal phenotype of the original abp1-1 and abp1-1s alleles can be complemented by the functional copy of the BSM gene that is present in the new abp1-c1 and abp1-TD1 alleles, but not by the bsm loss-of-function or the abp1-1 mutants themselves. Therefore, the originally described abp1 mutants are, indeed, loss-of-function alleles of the BSM gene and at least abp1-1 is a double loss-of-function mutant of ABP1 and BSM. Thus, the embryo-lethal phenotype previously attributed to the abp1 loss-of-function alleles is a result of the mutation in the neighboring BSM gene. For this reason, we also propose to re-annotate the abp1-1 and abp1-1s alleles as abp1-1/bsm1-2 and bsm1-3 (for this line it remains unclear if the insertion disrupts both ABP1 and BSM expression), respectively.
The abp1 and bsm allelic test together with no embryonic defects in the true abp1 knock-out lines makes it clear that no role has been identified for ABP1 in early embryogenesis. However, this clarification of abp1 knock-out genotypes does not per se undermine all experimentation with ABP1 genetic tools. The other ABP1 genotypes comprise conditional and constitutive gain-of-function alleles, two types of conditional knock-downs, as well as a weak abp1-5 point mutation (Braun et al., 2008; Covanova et al., 2013; Grones et al., 2015; Robert et al., 2010; Tromas et al., 2009; Xu et al., 2010). In several analyzed cellular processes including auxin-dependent ROP-GTPase activation, auxin-regulated endocytosis, E3 ligase-mediated ubiquitination, or cortical microtubule reorientation (Robert et al., 2010; Sassi et al., 2014; Tromas et al., 2013; Xu et al., 2010; Xu et al., 2014) this genetic material produced fully internally consistent data sets. Furthermore, loss-of-function mutants in TMK receptor-like protein kinases, which interact with ABP1 in an auxin-inducible manner, show largely overlapping phenotypes with abp1 mutants (Xu et al., 2014). Altogether, these data support the importance of the ABP1/TMK auxin perception and signaling mechanism in plant development and physiology. One possible explanation for the absence of obvious phenotype defects in true abp1 knock-outs is a presence of another copy of the ABP1 gene in the genome of A. thaliana that could escape the gene annotation during genome assembly and that could stay undetected by Southern blot analysis if the two copies were located close together. However, by in silico analysis of the ABP1 locus coverage we excluded this possibility. Other possible ways to reconcile the absent phenotype defects in the knock-outs with observations from the other genetic material include a genetic compensation mechanism that can be triggered by independent deleterious loss-of-function mutations but not by the genetic knock-downs of the same genes as shown for example in zebrafish (Rossi et al., 2015). Such compensation machinery could involve previously largely overlooked ABP1-like proteins from the germin family that are not a close sequence homologs of ABP1 but share some common characteristic features and some of them have been identified by their binding to auxin (reviewed in Napier et al., 2002).
Importantly, with the controversy of the embryonic phenotypes in different abp1 mutant alleles clarified, we can move on and with the updated genetic toolbox clarify the role of ABP1 in auxin signaling and plant development. The high affinity binding of ABP1 to auxin, universal occurrence of the ABP1 genes in the genomes from algae to higher plants and its highly conserved structure argue for its importance throughout the plant kingdom and promise further interesting discoveries.
F1000Research: Dataset 1. Dataset 1, 10.5256/f1000research.7143.d104552
F1000Research: Dataset 2. Dataset 2, 10.5256/f1000research.7143.d104553
F1000Research: Dataset 3. Dataset 3, 10.5256/f1000research.7143.d104554
JF and JM designed the experiments and wrote the manuscript, JM performed all experiments and analyzed the data except the short read coverage experiment, MD performed the genome coverage experiment, analyzed the data and helped with writing the manuscript, TB helped with planning and evaluating the genome coverage experiment. All authors have seen and agreed to the final content of the manuscript.
This work was supported by ERC Independent Research grant (ERC-2011-StG-20101109-PSDP to JF). JM internship was supported by the grant “Action Austria – Slovakia”.
We would like to thank Elena Babyichuk and Sergei Kushnir for providing bsm1-1 mutant seeds. We also like to thank Stephane Rombauts for useful advices for genome coverage analysis. We are thankful to Daniel von Wangenheim for final processing of the images.
Allelic test between A. thaliana bsm and abp1 knock-out mutants. Complete table with number of scored seeds for all crosses made. The percentage of white seeds from the total number of scored seeds is shown in brackets for each tested combination. N/A = data not available,♀ = plant was used as a pollen recipient, ♂ = plant was used as a pollen donor.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 23 Oct 15 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)