Keywords
Cannabis, Microbiome, Mycotoxins, Cannabidiol, Paxilline, Citrinin, qPCR, Culture, Next generation sequencing
This article is included in the Agriculture, Food and Nutrition gateway.
Cannabis, Microbiome, Mycotoxins, Cannabidiol, Paxilline, Citrinin, qPCR, Culture, Next generation sequencing
Many states in the U.S. are crafting regulations for microbial detection on Cannabis in absence of any comprehensive survey of Cannabis microbiomes. A few of these regulations are inducing growers to “heat kill” or pasteurize Cannabis flowers to lower microbial content. While this is a harmless suggestion, we must remain aware of how these drying techniques often create false negatives in culture-based safety tests used to monitor colony-forming units (CFU). Even though pasteurization may be effective at sterilizing some of the microbial content, it does not eliminate various pathogenic toxins or spores. Aspergillus spores and mycotoxins are known to resist pasteurization1,2. Similar thermal resistance has been reported for E. coli produced Shiga toxin3. While pasteurization may reduce CFU’s used in petri-dish or plating based safety tests, it does not reduce the microbial toxins, spores or DNA encoding these toxins.
Mycotoxin monitoring in Cannabis preparations is important since aflatoxin produced by Aspergillus species is a carcinogen. The clearance of aflatoxin requires the human liver enzyme CYP3A4 and this liver enzyme is potently inhibited by cannabinoids4,5. Modern day Cannabis flowers can produce up to 25% (w/v) cannabinoids presenting potent inhibition of CYP3A4 and CYP2C19. Health compromised patients exposed to aflatoxin and clearance-inhibiting cannabinoids raise new questions in regards to the current safety tolerances to aflatoxin. Similarly, Fusarium species are known to produce fungal toxins and has proven to be difficult to selectively culture with tailored media6–8. This is a common fault of culture-based systems as carbon sources are not exclusive to certain microbes and only 1% of microbial species are believed to be culturable9.
While these risks have been well studied in the food markets, the presence of the microbial populations present on Cannabis flowers has never been surveyed with next generation sequencing techniques10–15. With the publication of the Cannabis genome16,17 and many other pathogenic microbial genomes, quantitative PCR assays have been developed that can accurately quantify fungal DNA present in Cannabis samples18. Here, we analyze the yeast and mold species present in 10 real world, dispensary-derived Cannabis samples by quantitative PCR and sequencing, and demonstrate the presence of several mycotoxin producing fungal strains that are not detected by widely used culture-based assays.
3.55ml of tryptic soy broth (TSB) was used to wet 250mg of homogenized flower in a whirlpack bag. TSB was aspirated from the reverse side of the 100μm mesh filter and placed into a BiolumixTM growth vial and spread onto a 3M Petri FilmTM and a SimPlateTM (3M PetrifilmTM 3M Microbiology, St. Paul, MN, USA; SimPlatesTM Biocontrol Systems, Bellevue, WA, USA; BioLumixTM Neogen, Lansing MI, USA) according to the respective manufacturers’ recommendations. BiolumixTM vials were grown and monitored for 48 hours while Petri-filmsTM and SimPlatesTM were grown for 5 days. Petri-filmsTM and SimPlatesTM were colony counted manually by three independent observers. Samples were tested on total coliform, total entero, total aerobic, and total yeast and mold. Only total yeast and mold discrepancies were graduated to sequencing.
Plant DNA was extracted with SenSATIVAx according to manufacturers’ instructions (Medicinal Genomics part #420001). DNA was eluted with 50μl ddH20.
PCR was performed using 5μl of DNA (3ng/μl) 12.5μl 2X LongAmp (NEB) with 1.25μl of each 10μM MGC-ITS3 and MGC-ITS3 primer (MGC-ITS3; TACACGACGTTGTAAAACGACGCATCGATGAAGAACGCAGC) and (MGC-ITS3R; AGGATAACAATTTCACACAGGATTTGAGCTCTTGCCGCTTCA) with 10μl ddH20 for a 25μl total reaction. An initial 95°C 5 minute denaturization was performed followed by 40 cycles of 95°C for 15s and 65°C for 90s. Samples were purified with 75μl SenSATIVAx, washed twice with 100μl 70% EtOH and bench dried for 5 minutes at room temperature. Samples were eluted in 25μl ddH20.
DNA libraries were constructed with 250ng DNA using NEB’s NEBNext Quick ligation module (NEB # E6056S). End repair used 3μl of enzyme mix, 6.5μl of reagent mix, 55.5μl of DNA + ddH20. Reaction was incubated at 30°C for 20 minutes. After end repair, ligation was performed directly with 15μl of blunt end TA mix, 2.5μl of Illumina adaptor (10μM) and 1μl of ligation enhancer (assumed to be 20% PEG 6000). After 15 minute ligation at 25°C, 3μl of USER enzyme was added to digest the hairpin adaptors and prepare for PCR. The USER enzyme was tip-mixed and incubated at 37°C for 20 minutes. After USER digestion, 86.5μl of SenSATIVAx was added and mixed. The samples were placed on a magnet for 15 minutes until the beads cleared and the supernatant could be removed. Beads were washed twice with 150μl of 70% EtOH. Beads were left for 10 minute to air dry and then eluted in 25μl of 10mM Tris-HCl.
25μl 2X Q5 polymerase was added to 23μl of DNA with 1μl of i7 index primer (25μM) and 1μl universal primer (25μm). After an initial 95°C for 10s, the library was amplified for 15 cycles of 95°C 10s, 65°C 90s. Samples were purified by mixing 75μl of SenSATIVAx into the PCR reaction. The samples were placed on a magnet for 15 minutes until the beads cleared and the supernatant could be removed. Beads were washed twice with 150μl of 70% EtOH. Beads were left for 10 minute to air dry and then eluted in 25μl of 10mM Tris-HCl. Samples were prepared for sequencing on the MiSeq version 2 chemistry according to the manufacturers’ instructions. 2×250bp reads were selected to obtain maximal ITS sequence information.
Primers described by Shirazi-zand et al. were utilized to amplify a segment of the 725bp PaxP gene. 25μl LongAmp (NEB) 4μl 10μM primer, 1μl DNA (14ng/μl), 20μl ddH20 to make a 50μl PCR reaction. Cycling conditions were slightly modified to accommodate a different polymerase. 95°C for 30s followed by 28 cycles of 95°C 15s, 55°C for 30s, 65°C 2.5 minutes. Samples were purified with 50μl of SenSATIVAx as described above. 1μl of purified PCR product was sized on Agilent HS 2000 chip. Nextera libraries and sequencing were performed according to instructions from Illumina using 2×75bp sequencing on a version 2 MiSeq.
Citrinum forward GATTTTCCAAAATGCCGTCT and Citrinum reverse GCTCAAGCATTAATCTAGCTA primers were used with identical PCR conditions as above with the exception using 35 cycles of PCR. Samples were purified with 50μl of SenSATIVAx as described above. 1μl of purified PCR product was sized on Agilent HS 2000 chip. Nextera libraries and sequencing were performed according to instructions from Illumina using 2×75bp sequencing on a version 2 MiSeq. Reads were mapped to Genbank accession number LKUP01000000. Mappings were confirmed using BLAST to NCBI to ensure the strongest hits were to P. citrinum.
Reads were demultiplexed and trimmed with Casava 1.8.2 and trim_galore v0.4.1 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). FLASH v1.2.1119 was used to merge the reads using max_overlap 150. The reads were aligned to microbial references using MG-RAST v3.220. Alignments and classifications were confirmed with a second software tool from One Codex (https://onecodex.com/) and critical pathways identified for further evaluation with PCR of toxin producing genes. Reads are deposited in NCBI under SRA accession: SRP065410. Nextera 2×75bp sequencing of the PaxP gene was mapped to accession number HM171111.1 with CLCbio Workstation V4 at 98% identity over 80% of the read. One Codex analysis was put into Public mode under the following public URLs:
Australian Bastard:
https://app.onecodex.com/analysis/public/201e7f1642e04a3c
https://app.onecodex.com/analysis/public/58f1e03c10434bfa
KD4:
https://app.onecodex.com/analysis/public/2e86e262817246c4
https://app.onecodex.com/analysis/public/1abd5b60446140a0
KD6:
https://app.onecodex.com/analysis/public/a92d3dff5485499d
https://app.onecodex.com/analysis/public/8d72e2514e564ecd
KD8:
https://app.onecodex.com/analysis/public/8d72e2514e564ecd
https://app.onecodex.com/analysis/public/d6e2e0bcfba3469f
Liberty Haze:
https://app.onecodex.com/analysis/public/7bcd650fa5544f2c
https://app.onecodex.com/analysis/public/7f0feb6cb0a94d56
Girls Scout Cookie:
https://app.onecodex.com/analysis/public/a71b1ce8331c461d
https://app.onecodex.com/analysis/public/8d6f10c7ee684f93
Jakes Grape:
https://app.onecodex.com/analysis/public/bc8af5ed19e5407a
https://app.onecodex.com/analysis/public/99d7a4a2f7af486b
RECON:
https://app.onecodex.com/analysis/public/8a22a16cc2e24731
https://app.onecodex.com/analysis/public/0af6ae26a01f48d5
GreenCrack:
https://app.onecodex.com/analysis/public/6114843d2eb3425e
https://app.onecodex.com/analysis/public/3eee642786c54a88
LA Confidential:
https://app.onecodex.com/analysis/public/01e8aefb0d4f4f62
https://app.onecodex.com/analysis/public/b74c2988fcd84e38
NYC Diesel:
A commercially available total yeast and mold qPCR assay (TYM-PathogINDICAtor, Medicinal Genomics, Woburn MA) was used to screen for fungal DNA in a background of host Cannabis DNA. The TYM qPCR assay targets the ribosomal DNA Internal Transcribed Spacer region 2 (ITS2) using modified primers described previously21,22. Fungal DNA amplified using these primers may also be subjected to next generation sequencing to identify the contributing yeast and mold species. ITS sequencing has been widely used to identify and enumerate fungal species present in a given sample23.
We purified DNA from Cannabis samples obtained from two different geographic regions (Amsterdam and Massachusetts) several years apart (2011 and 2015). The majority of samples purified and screened with ITS qPCR were negative for amplification signal implying reagents clean of fungal contamination. Six of the 17 dispensary-derived Cannabis samples tested positive for yeast and mold in the TYM qPCR assay. These results were compared with the results derived from three commercially available culture-based detection systems for each of the 17 samples (3M PetrifilmTM 3M Microbiology, St. Paul, MN, USA; SimPlatesTM Biocontrol Systems, Bellevue, WA, USA; BioLumixTM Neogen, Lansing MI, USA; Figure 1). Of the 6 qPCR positive samples, two tested negative in all 3 culture-based assays and four tested negative in 1 or 2 of the culture-based assays (Table 1). None of the qPCR negative samples tested positive in any of the culture-based assays. Each of the 6 discordant samples was subjected to ITS sequencing to precisely identify the collection of microbes present. Four additional samples from a different geographic origin (Amsterdam) were also subjected to ITS sequencing, for a total of 10 Cannabis samples.
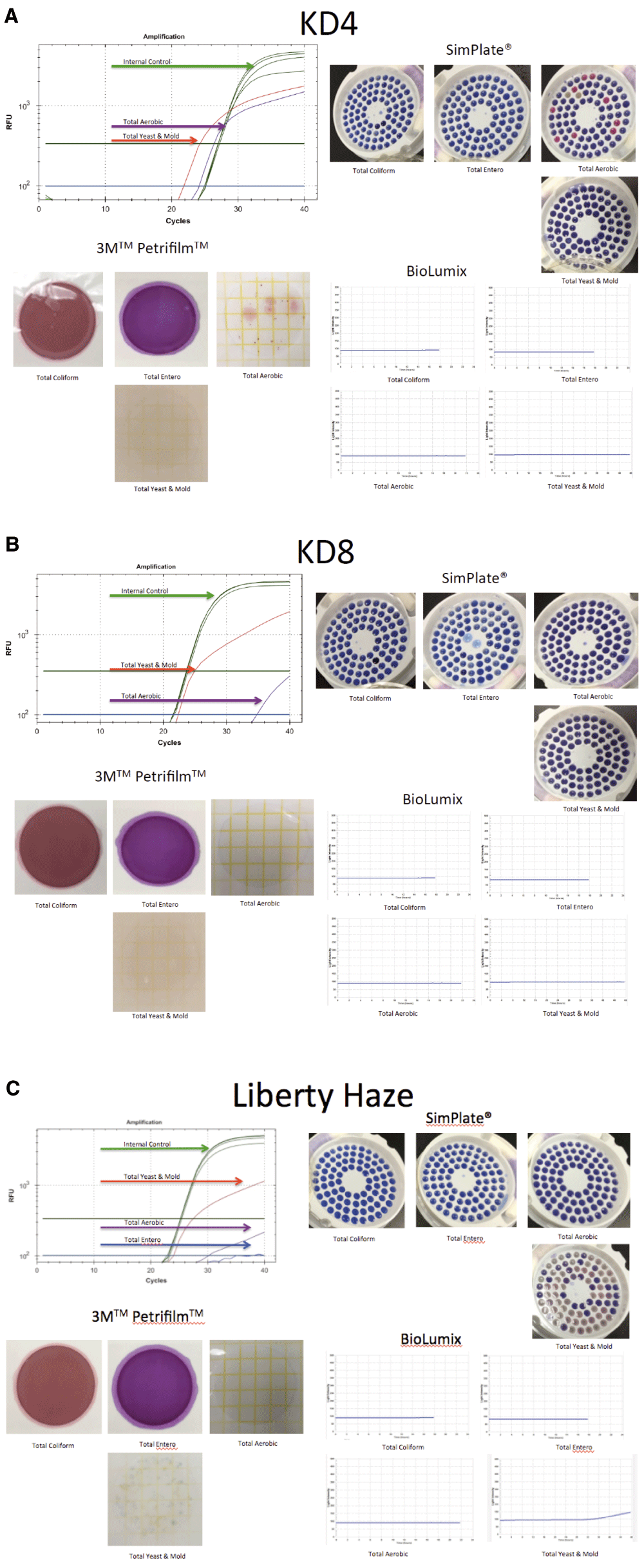
Figure 1A. qPCR signal from TYM (red line) test run concurrently (multiplexed) with a plant internal control marker (green line). This marker targets a conserved region in the Cannabis genome and should show up in every assay (upper left). SimPlates count the number of discolored wells (purple to pink) as a proxy for CFU/gram. Only total aerobic show growth (upper right). Petrifilm only demonstrate colonies on total aerobic platings (lower left). Biolumix demonstrate no signal across all 4 tests (lower right). Figure 1B. Sample KD8 fails to culture any total yeast and mold yet demonstrates significant TYM qPCR signal. Sample was graduated to ITS based next generation sequencing. Figure 1C. Sample Liberty Haze was tested with 3 culture based methods and compared to qPCR. Sample was graduated to ITS based next generation sequencing.
Biolumix had the lowest sensitivity failing to pick up 4/17 samples detected with other culture-based platforms. qPCR identified 2 samples that were not picked up by any other method. Positive qPCR samples were sequenced to identify the contributing signal. Highlighted samples fail the 10,000 CFU/g cutoffs which equates to a Cq of 26 on the qPCR assay according to the manufacturers’ instructions. (f) is fail or over 10,000 CFU/g. (p) is pass or under 10,000 CFU/g. The raw CFU numbers can be deduced by dividing the CFU number by the 1,000 fold dilution factor used in this study.
Each discordant sample presented with an array of microbial species, as shown in Figure 2. No sample presented with a single dominant species, and each sample displayed multiple species of interest. Of particular concern were the identified DNA sequences from toxin producing species: Aspergillus versicolor24–28, Aspergillus terreus29, Penicillium citrinum30–32, Penicillium paxilli33,34.
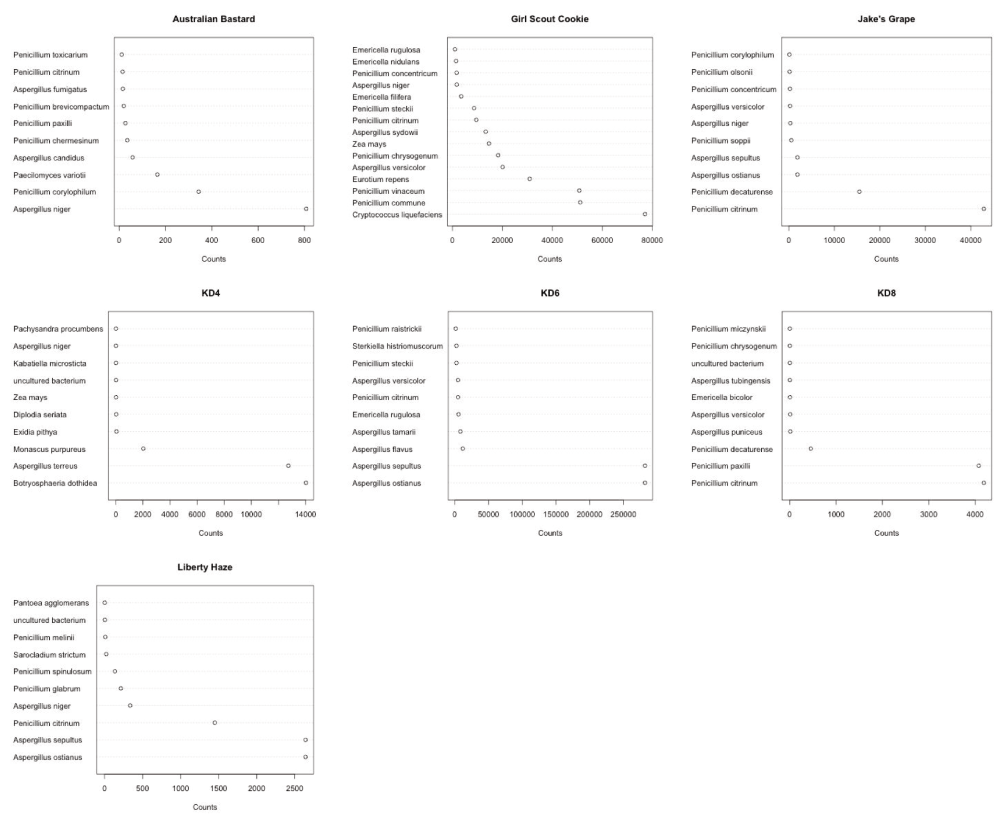
Penicillium and Aspergillus are commonly found (Y axis) but at different read counts in each sample (X axis). Read counts are more a reflection of sample normalization for sequencing than inter sample quantitation provided by qPCR.
We further analyzed the ITS sequence alignments using the whole genome shotgun based microbiome classification software known as One Codex35. Nine of the ten samples sequenced showed the presence of P. paxilli (Figure 3). To verify the accuracy of this ITS phylotyping, a gene involved in the paxilline toxin biosynthesis pathway of P. paxilli was amplified with PaxPss1 and PaxPss2 primers described by Saikia et al.36 The resulting 725bp amplicon (expected size) was sequenced to confirm the presence of the P. paxilli biosynthesis gene in the Cannabis sample KD8 (Figure 4). This was successfully repeated with primers designed to target genes in the citrinin pathway of P. citrinum. There were some discrepancies between the results derived from the two software platforms (One Codex and MG-RAST). The MG-RAST analysis, using merged, paired reads correlated better with the PCR results. While One Codex predicted and confirmed KD8 as having the highest P. paxilli content, the One Codex platform is optimized for whole genome shotgun data and may not be able to differentiate the 18S sequence differences (391/412 aligned bases) between these two species with a K-mer based approach.
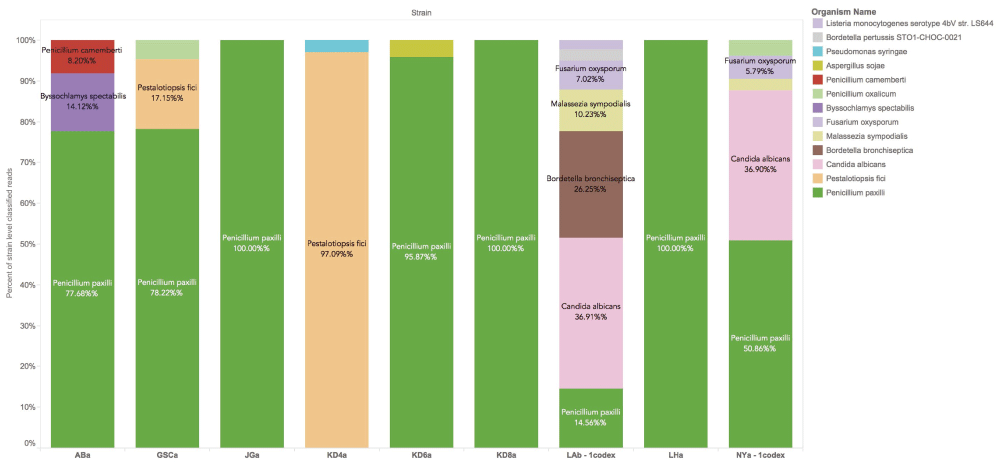
P. paxilli is the most frequently found contaminant in Cannabis flowers. P. citrinum is not in the One Codex database at this time. One Codex utilizes a fast k-mer based approach for whole genome shotgun classification and can be influenced by read trimming and database content. The reads provided to MG-RAST were trimmed and FLASH’d (paired end reads merged when overlapping) prior to classification. K-mer based approaches can significantly differ from longer word size methods and this underscores the importance of confirmatory PCR in microbiome analysis.
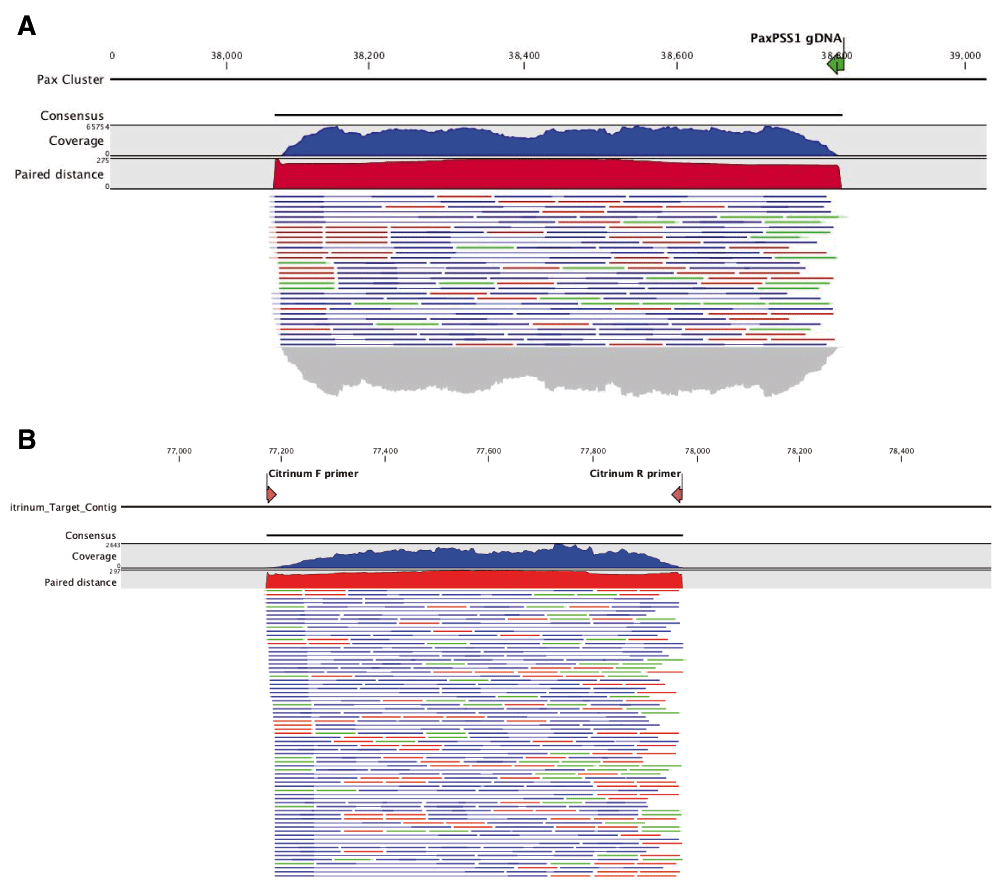
Citrinum primers we designed from Genbank accession number LKUP01000753. Paxilline primers were used as described in Saikia et al. PCR products were made into shotgun libraries with Nextera and sequenced on an Illumina MiSeq with 2×75bp reads to over 10,000X coverage. Reads were mapped with CLCbio 4 to NCBI accession number HM171111.1 (A) and LKUP01000000 respectively (B). Paired reads are displayed as blue lines, green and red lines are unpaired reads. Read coverage over the amplicons are depicted in a blue histogram over the cluster while paired end read distance is measured in a red histogram over the region. Off target read mapping is limited. P. paxilli mappings are displayed on top (A) and P. citrinum mappings are displayed on bottom (B). Alignment of PCR primers to P. paxilli reference shows a 5 prime mismatch that is a result of the primers being designed to target spliced RNA according to Saikia et al.
With the confirmed presence of P. paxilli, we are curious to find out whether the toxin, paxilline, is present in the samples. Development of monoclonal antibodies to paxilline has recently been described37, but commercial ELISA assays with sensitivity under 50ppb do not appear to be available at this time. A >50ppb multiplexed ELISA assay is available from Randox Food Diagnostics (Crumlin, UK). Detection with LC-MS/MS has also been described38,39, however, and experiments are underway to determine whether paxilline can be identified in the background of cannabinoids and terpenes present in Cannabis samples.
Several potentially harmful fungal species were detected in dispensary-derived Cannabis samples by qPCR and subsequent sequencing in this study. Three different culture-based assays failed to detect all of the positive samples and one, BioLumixTM, detected only one out of 7 positive samples. A review of the literature suggests that Penicillium microbes can be cultured on CYA media, but some may require colder temperatures (21-24C) and 7 day growth times40. Of the Penicillium, only P. citrinum has been previously reported to culture with 3M Petri-Film41. In addition, several studies have demonstrated plant phytochemicals and terpenoids like eugenol can inhibit the growth of fungi42. It is possible the different water activity of the culture assay compared to the natural terpene rich flower environment is contributing to the false negative test results.
Quantitative PCR is agnostic to water activity and can be performed in hours instead of days. The specificity and sensitivity provides important information on samples that present risks invisible to culture based systems. The draw back to qPCR is the method’s indifference to living or non-living DNA. While techniques exist to perform live-dead qPCR, the live status of the microbes is unrelated to toxin potentially produced while the microbes were alive. ELISA assays exist to screen for some toxins43. Current state-recommended ELISA’s do not detect citrinin or paxilline, the toxins produced by P. citrinum and P. paxilli, respectively. The predominance of these Penicillium species in a majority of the samples tested is interesting. Several Penicillium species are known to be endophytes on various plant species, including P. citrinum10, and this raises the question of whether they are also Cannabis endophytes.
Paxilline is a tremorgenic and ataxic potassium channel blocker and has been shown to attenuate the anti-seizure properties of cannabidiol in certain mouse models44–46. Paxilline is reported to have tremorgenic effects at nanomolar concentrations and is responsible for Ryegrass-staggers disease47. Cannabidiol is often used at micromolar concentrations for seizure reduction implying sub-percentage contamination of paxilline could still be a concern. Citrinin is a mycotoxin that disrupts Ca2+ efflux in the mitochondrial permeability transition pore (mPTP)48–55. Ryan et al. demonstrated that cannabidiol affects this pathway suggesting a potential concern for CBD-mycotoxin interaction56. Considering the hydrophobicity of paxilline and the recent interest in the use of cannabidiol derived from Cannabis flower oils for drug resistant epilepsy, more precise molecular screening of fungal toxins may be warranted57–62.
Our survey of Cannabis flowers in this study was limited. Further screening will be required to define a set of tests that can adequately capture all risks. While ELISA assays are easy point of use tests that can be used to detect fungal toxins, they can suffer from lack of sensitivity and cross reactivity. ITS amplification and sequencing offers hypothesis-free testing that can complement the lack of specificity in ELISA assays. Appropriate primer design can survey a broad spectrum of microbial genomes while affording rapid iteration of design. Quantitative PCR has also demonstrated single molecule sensitivity and linear dynamic range over 5 orders of magnitude offering a very robust approach for detection of microbial risks. This may be important for the detection of nanomolar potency mycotoxins. Further studies are required to validate better detection methods for these toxins and verify whether paxilline or citrinin are present on Cannabis at concentrations that present a clinical risk.
These results demonstrate that culture based techniques superimposed from the food industry should be re-evaluated based on the known microbiome of actual Cannabis flowers in circulation at dispensaries. Several mycotoxin producing molds were detected that can potentially interfere with the medical use of cannabidiol. These microbes failed to grow on traditional culture-based platforms but were rapidly detected with molecular based techniques. Further studies are required to quantitate the presence and concentration of mycotoxin production.
KJM designed the study and performed the One-Codex analysis and PCR verifications.
JS designed and ran the culture and qPCR laboratory experiments.
LZ assisted in the figure generation and laboratory experiment.
YH assisted in sequencing and PCR confirmation of Pax.
VT- read alignment, MG-RAST, primer design and analysis.
TF- Sample tracking software, figure generations, ITS software comparisons.
DS- Manuscript construction and review.
The authors are employees of Medicinal Genomics Corporation (MGC). MGC manufactures qPCR reagents utilized in this study.
John McPartland, Cindy Orser, Brad Douglass, Joost Heeroma, Nick Greenfield, Rebecca McKernan and Kellie Dodd for thoughtful advice.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
References
1. Cao HX, Schmutzer T, Scholz U, Pecinka A, et al.: Metatranscriptome analysis reveals host-microbiome interactions in traps of carnivorous Genlisea species.Front Microbiol. 2015; 6: 526 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 10 May 16 |
read | |
|
Version 1 10 Dec 15 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)