Keywords
multi-scale modeling, JSim, systems biology, physiological modeling, reproducibility, uncertainty quantification
multi-scale modeling, JSim, systems biology, physiological modeling, reproducibility, uncertainty quantification
Many attempts have been made to provide modular programming systems for physiological applications (Erson et al., 2012; Krause et al., 2010; Mirschel et al., 2009; Smith et al., 2009). We describe our system as a semi-automated modular programming construction. It is simple and not conceptually novel, but is easy to learn and use. For developing a series of models of increasing complexity, Modular Program Constructor (MPC) can serve well as the basis of the modeling code. The perspective is to take a modular approach; this means that for multi-scale modeling one builds from simple elements initially and then uses multi-modular constructs as modules in higher level systems.
In MPC, a module can be any set of variable declarations, parameter declarations and mathematical equations that represent a process. This broad definition of a module has a broad variety of applications: from a simple first order enzyme reaction, to a complete model of coronary blood flow through heart muscle, which can then be incorporated into a yet larger systemic model.
Based on ModelBuilder, which used FORTRAN to parse code and define directives (Raymond, 2008), we designed the current version in Java and renamed it MPC. It is now refined, simplified, robust, and used to develop several new applications (Raymond et al., 2011; Raymond et al., 2012). The models include time-dependent two-dimensional models in both Cartesian and cylindrical coordinates (Raymond et al., 2011; Raymond et al., 2012), as well as whole organ models with heterogeneous flow re-implementing earlier complex whole organ models of substrate metabolism (Bassingthwaighte et al., 1989).
MPC is a pre-compiler written in Java. It reads a text input file, parses the file for directives, and generates a text output file based on those directives. MPC is built upon the Mathematical Modeling Language (MML) of JSim (http://www.physiome.org/jsim/) (Butterworth et al., 2014). It has been designed to work with JSim's MML and currently requires JSim to run the model output file that MPC produces. Through JSim, the final constructed model can be exported into Systems Biology Markup Language (SBML, http://sbml.org/Main_Page), CellML (https://www.cellml.org/), and downloaded from these sources to other simulation platforms (Smith et al., 2014). MPC currently is executed as command line utility and requires the Java runtime environment (https://java.com/).
MPC has three components:
1. MML, the mathematical modeling language of JSim, is a declarative language designed for solving all the equations simultaneously; it is not procedural. MML is used for declaring parameters and variables, for defining algebraic equations, ordinary differential equations, and partial differential equations with their associated constraints, and initial and boundary conditions.
2. Modules, are sets of MML code libraries which are variable declarations, parameter declarations, or mathematical equations for a particular process, for example, flow along a capillary, diffusion within a region, a chemical reaction, transport across a membrane, or even a whole organ. These are archived, forming libraries of operational code. This allows the user to generate multi-scale models with different sub-models to use in testing a hypothesis against data, i.e. validity testing. For example, there have been a variety of models developed to describe the transmembrane sodium pump, NaKATPase which uses ATP to pump sodium out of, and potassium into, the cell. All of these models have the same essential external influences: the Na and K ion concentrations and the transmembrane electrical potential. Having a library of the MML code for the variant modules allows one to insert one's choice quickly into the template for the cell model. Changing combination(s) rapidly to match solutions with experimental results is invaluable for the early phases of developing alternative hypotheses.
3. Directives, the third component, comprises the set of instructions used by the MPC program to select processes and gather the code from existing modules, renaming parameters and variables to reflect the new purposes for which they will function, and automatically combining the mathematical structures into new structures. The directives control the identification, fetching and relabeling of variables and parameters, and the assembly and recombination of code into new equations. All MPC directives start with '//%'.
The MPC input file guides the construction of a model made of previously existing modules. It combines MML with “directives” embedded as comments. It uses code from other JSim model files that have been annotated so that they can be read by MPC, yet without interfering with their operability. MPC may also combine models with other models or with modules of preconstructed code from libraries. These modules are specified within a library with the START and END directive. A library with a few elementary operators from which we will build a model in out next step is illustrated below:
CodeLibrary.mod:
//------------------------- ODE DOMAINS
//%START odeDomains // START...END directives used to specify a module.
realDomain t s; t.min=0; t.max=16; t.delta = 0.1;
//%END odeDomains
//------------------------- flowCalc
//%START flowCalc
C:t = (F/V)*(Cin-C);
//%END flowCalc
//------------------------- EXCHANGE CACULATIONS
//%START exchangeCalc
C1:t = PS/V1*(C2-C1); // Exchange between two compartments
C2:t = PS/V2*(C1-C2);
//%END exchangeCalc
//------------------------- REACTION A->B
//%START reactionCalc
real G = 5 ml/(g*min); // Const reaction rate.
A:t = -G/V*A;
B:t = G/V*A;
//%END reactionCalc
//------------------------- MM REACTION A->B
//%START MMreactionCalc
real KmA =1.0 mM, VmaxA =2 umol/(g*min); // MM constant and max velocity of rxn
real G(t) ml/(g*min); // MM reaction rate
G = (VmaxA/(KmA+A));
A:t = -G*(A)/V;
B:t = G*(A)/V;
//%END MmreactionCalcIn JSim's MML, the colon signifies the derivative: C:t means dC/dt. Within MPC we can write MML code directly or import code from operational JSim models that have been annotated to identify components. An example is a three species (A, B, C), two compartment model with two reactions in compartment two (Figure 1) with species concentrations described by ordinary differential equations (ODE). Species A enters, with flow F, a compartment with volume V1 and passive exchange between a second compartment with volume V2, where A reacts at rate GA2B to form B and B reacts with C at rate GB2C, a Michaelis-Menten reaction.

Ain is A entering compartment 1 with flow F (No flow in for species B, C). Passive exchange between compartments for all three species and reactions only occur in compartment 2.
The MPC file defines the domain, parameters, variables, and initial conditions first. Using directives listed in ‘Example.mpc’, model code is extracted from the file ‘CodeLibrary.mod’ shown above. Values and variable names needing replacement throughout the final model are specified by the REPLACE directive along with the '%symbol%' placeholder. The use of the REPLACE, GET, COLLECT, INSERTSTART and INSERTEND directives are used in Example.mpc shown below:
Example.mpc:
//%REPLACE %CL% =("CodeLibrary.mod") // Library to get code from, replace all
// occurrences of %CL% with CodeLibrary.mod
//%REPLACE (%N%=(“1”,”2”), %vol%=("0.05","0.05")) // Two compartments with volumes, replace
// all occurrences of %N% with 1,2 and %vol% with 0.05, 0.15
//%REPLACE (%AB%=("A","B","C") %PS3%=("6","5","4")) // 3 species, PS init values.
import nsrunit; unit conversion on; // Use SI units for this model.
math example { // model declaration
// INDEPENDENT VARIABLES
//%GET %CL% odeDomains() // Get odeDomains section from CodeLibrary.mod
//%INSERTSTART a2bParmsVars // Specify params and vars section
// PARAMETERS
real Flow = 1 ml/(g*min); // Flow rate
real PS%AB%12 = %PS3% ml/(g*min); // Conductances: PSA12,PSB12,PSC12
real V%N% = %vol% ml/g; // Volume of V1, V2
extern real %AB%in(t) mM; // Inflowing concentrations
// DEPENDENT VARIABLES
real %AB%%N%(t) mM; // A1,A2,B1,B2,C1,C2
// INITIAL CONDITIONS (IC's)
when(t=t.min) %AB%%N%=0; // Defines IC's for the ODEs
//%INSERTEND a2bParmsVars // End params and var sec
//%INSERTSTART a2bCalc // Specify calc section
// ODE CALCULATIONS
//%GET %CL% reactionCalc ("A=A2","B=B2","V=V2","G=Ga2b") // A->B reaction
//%GET %CL% MMreactionCalc ("A=B2","B=C2","V=V2","G=Gb2c", // B ->C MM reaction
//% "KmA=KmB2",“VmaxA = VmaxB2", "KmA = KmB2") // B ->C MM reaction continued
//%GET %CL% flowCalc ("Cin=%AB%in","C=%AB%1","V=V1","F=Flow","D=D%AB%1")
//%GET %CL% exchangeCalc ("C1=%AB%1","PS=PS%AB%12","C2=%AB%2")
//%COLLECT("%AB%%N%:t") //Group all ODE calculations for a species together
//%INSERTEND a2bCalc
} // curly bracket ends modelThe GET directive warrants further explanation: it identifies a code library file and module name within the library to insert into the model, and changes old names (names of parameters and variables in the module) to new model names. From the example above, //%GET %CL% reactionCalc ("A=A2","B=B2","V=V2","G=Ga2b") will get the module named 'reactionCalc' in file 'CodeLibrary.mod' and replace the variable names with the new model names ("A=A2", etc).
The MPC directives control the identification, fetching, relabeling of variables and parameters, and assembling and recombining code into new equations. The directives extract equations from files, changing the names of the module variables to application specific names and assemble the code into combined equations. The code resulting from these instructions provides a complete program (Example.mod), with no further intervention on the part of the user except to adjust parameters, solution time step length and set up graphics in JSim to display solutions, as shown in Figure 2.
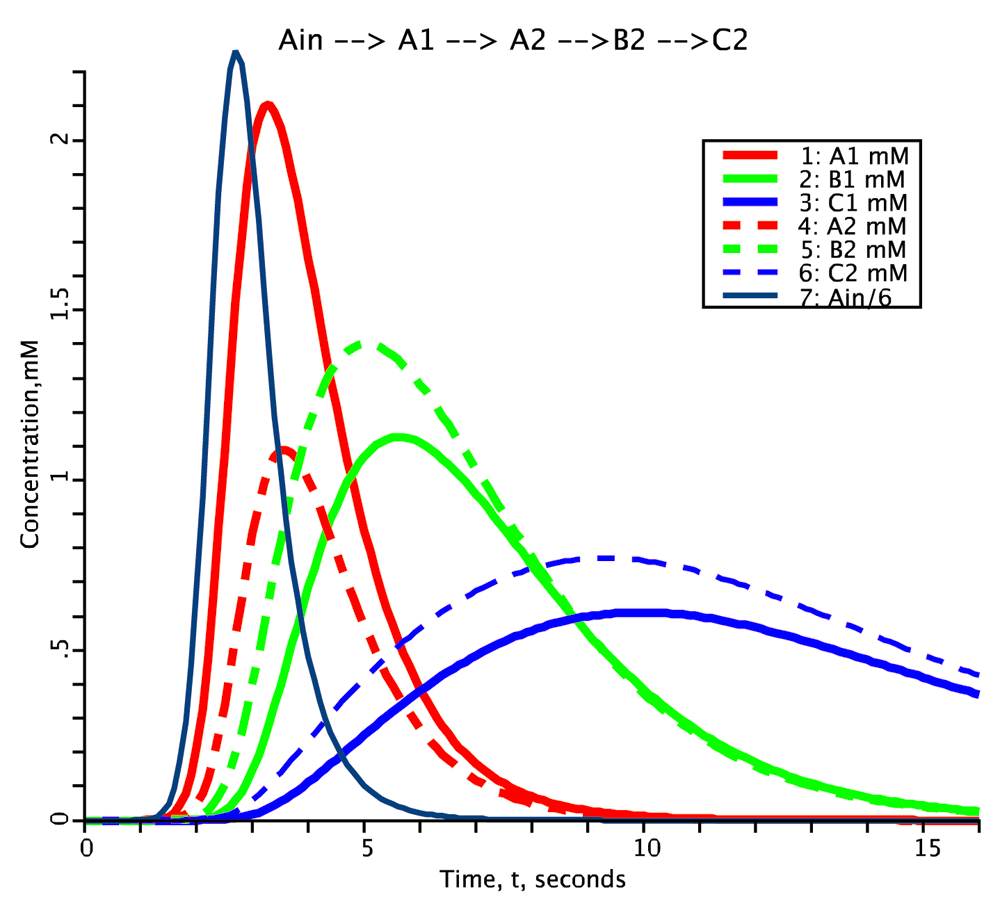
Species concentrations plotted as a function of time. Species A (red), B (green), C (blue). Compartment 1: solid line, Compartment 2: dashed line. External input for species A is Ain, the black solid line.
MPC Output - Example.mod:
import nsrunit; unit conversion on; // Use cgs units
math example { // model declaration
// INDEPENDENT VARIABLES
realDomain t s; t.min=0; t.max=16; t.delta = 0.1;
//%START a2bParmsVars // Specify parameters and variables sect.
// PARAMETERS
real Flow = 1 ml/(g*min); // Flow rate
real PSA12 = 6 ml/(g*min); // Conductance
real PSB12 = 5 ml/(g*min); // Conductance
real PSC12 = 4 ml/(g*min); // Conductance
real V1 = 0.05 ml/g; // Volume
real V2 = 0.05 ml/g; // Volume
extern real Ain(t) mM; // Inflowing concentration
extern real Bin(t) mM; // Inflowing concentration, set to zero
extern real Cin(t) mM; // Inflowing concentration, set to zero
// DEPENDENT VARIABLES
real A1(t) mM; real A2(t) mM; real B1(t) mM;
real B2(t) mM; real C1(t) mM; real C2(t) mM;
// INITIAL CONDITIONS (IC's)
when(t=t.min) A1=0;
when(t=t.min) A2=0;
when(t=t.min) B1=0;
when(t=t.min) B2=0;
when(t=t.min) C1=0;
when(t=t.min) C2=0;
//%END a2bParmsVars // End parameters and variables section
//%START a2bCalc // Specify calculations section
real Ga2b = 5 ml/(g*min); // A ->B Const reaction rate.
real KmB2 =1.0 mM, VmaxB2 =2 umol/(g*min);// MM constant and max velocity of rxn
real Gb2c(t) ml/(g*min); // B ->C MM reaction rate
Gb2c = (VmaxB2/(KmB2+B2));
// ODE CALCULATIONS
A2:t = -Ga2b/V2*A2 +PSA12/V2*(A1-A2);
B2:t = Ga2b/V2*A2 -Gb2c*(B2)/V2 +PSB12/V2*(B1-B2);
C2:t = Gb2c*(B2)/V2 +PSC12/V2*(C1-C2);
A1:t = (Flow/V1)*(Ain-A1) +PSA12/V1*(A2-A1);
B1:t = (Flow/V1)*(Bin-B1) +PSB12/V1*(B2-B1);
C1:t = (Flow/V1)*(Cin-C1) +PSC12/V1*(C2-C1);
//%END a2bCalc
} // curly bracket ends modelThe process above is hardly worthwhile for small models but is highly efficient for larger models where flexibility in structure is desired. In the example above, converting the Ordinary Differential Equations (ODEs) to Partial Differential Equations (PDEs) requires a three line change. Addition of a new PDE e.g. for red blood cells in a capillary, takes four lines. For a five species, three region model, a three line change generates a 15 PDE model.
The small set of directives builds complex models from simple processes. MPC allows one to reliably reuse existing models in larger, multi-scale models. MPC encodes and preserves information about how a complex model is built from its modules allowing quick substitution of modules. The amount of actual code a user needs to write is reduced, especially for more complicated models. In MPC we have generated a full organ model with heterogeneity of flow, competitive transporters on the cell membranes, and reactions for multiple species (Bassingthwaighte et al., 2012) e.g. for adenosine processing in the heart. It is a 7-path, three region model that involves five species (adenosine, inosine, hypoxanthine, xanthine, and uric acid) in a sequential reaction chain. The model contains over 100 PDEs for convection, diffusion, and reactions.
Though a MPC-generated model is checked for syntax and unit balance through JSim, verification is required: analytical solutions can be written into the code to match specific limiting cases, but otherwise one depends on testing for mass, charge, or energy balances. Validation requires testing against data, independent of the construction method. These are key steps toward reproducibility and the VVUQ process. (VVUQ = verify, validate, uncertainty quantification; the latter defining predictive accuracy.) MPC as is, depends on semantic consistency throughout the libraries and models used. Automated systems using ontologies will help craft models (Gennari et al., 2011), but the great efficiency of MPC for construction begins to show when there are many modules in series/parallel arrangements as in biochemical networks or circulatory or airway mechanical modeling. UQ includes uncertainty in inputs and parameters, readily handled by JSim's Monte Carlo analysis, and in model structure. Structural uncertainty, a major challenge, defines a major role for MPC: inserting different choices from amongst similar but differently functioning modules, into a large, multi-modular model, and solving the system many times with the variant constituents illustrating uncertainty in the projected outcomes.
A limited set of directives in MPC, our Modular Program Constructor, allows us to build complex models using small models for simple processes. MPC encodes and preserves information about how a complex model is built from its modules allowing the researcher to quickly substitute or update modules to validate a hypothesis. The amount of actual code a user needs to write is reduced, especially for more complicated models.
Future updates will improve collection and insertion of model code, better identify external module 'connections' for easier incorporation into larger models, and more intelligent reconciliation of similar code between modules. The long-term strategy is to integrate MPC within JSim allowing the user to take advantage of JSim's MML compiler and graphical user interface to quickly merge code with less user intervention.
The Java code for MPC, the examples presented here, some more detailed examples, and instructions are available at http://www.physiome.org/software/MPC/.
MPC is released under a 3-clause ‘revised’ BSD license:
Copyright (C) 1999–2015 University of Washington
Developed by the National Simulation Resource
Department of Bioengineering, Box 355061
University of Washington, Seattle, WA 98195-5061.
Dr. J. B. Bassingthwaighte, Director
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
* Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer.
* Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.
* Neither the name of the University of Washington nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission.
MPC generated models for review at www.physiome.org are:
• Concentration profiles in capillary and tissue when exchange is diffusion-limited (http://www.physiome.org/jsim/models/webmodel/NSR/DiffusionLimitedProfiles/).
• ODE model of actin polymerization and depolymerization with tracking of bound nucleotide (http://www.physiome.org/jsim/models/webmodel/NSR/ActinCycle1/).
• Multiple tracer dilution estimates of D- and 2-deoxy-D-glucose uptake by the heart (http://www.physiome.org/jsim/models/webmodel/NSR/Kuikka1986BTEX30MP/).
Gary Raymond developed MPC. Bart Jardine currently maintains MPC source code and James Bassingthwaighte provides guidance and requirements for MPC development. All authors contributed to the design and organization of the paper and its writing and editing. All authors have seen and agreed to the final content of the manuscript.
Research has been supported by NIH grants HL088516 (J.B. Bassingthwaighte) and HL073598 (J.B. Bassingthwaighte), BE08417 (J.B. Bassingthwaighte), the Virtual Physiological Rat program GM094503 (PI: D.A. Beard), and the Cardiac Energy Grid HL199122 (PI: J.B. Bassingthwaighte). Grants supported whole group.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
References
1. Neal ML, Carlson BE, Thompson CT, James RC, et al.: Semantics-Based Composition of Integrated Cardiomyocyte Models Motivated by Real-World Use Cases.PLoS One. 2015; 10 (12): e0145621 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 3 (revision) 16 Jun 16 |
read | |
|
Version 2 (revision) 06 Apr 16 |
read | read |
|
Version 1 16 Dec 15 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)