Keywords
F1000Prime, Altmetrics, Mendeley, paper, evaluation
This article is included in the Research on Research, Policy & Culture gateway.
F1000Prime, Altmetrics, Mendeley, paper, evaluation
In this new version of the manuscript we have added a new Conclusions section, extended the literature review, methodological presentation and Discussion.
See the authors' detailed response to the review by Rodrigo Costas
See the authors' detailed response to the review by Stefanie Haustein
Interest in the broad impact of research (Bornmann, 2012, Bornmann, 2013, King’s College London and Digital Science, 2015) has resulted in new forms of impact measurements. Traditional forms of impact measurements using bibliometrics only allow the measurement of impact on research itself. These new forms which have been named as altmetrics (abbreviation of alternative metrics) pretend to measure the impact of research on other areas of society (than research) by counting the mentions of papers in social media: “Alternative metrics, sometimes shortened to just altmetrics, is an umbrella term covering new ways of approaching, measuring and providing evidence for impact” (Adie, 2014, p. 349). As altmetrics, the number of readers (on Mendeley), mirco-bloggers (on Twitter), and other consumers of research using social media are counted. Although scientometrics research on altmetrics is still in a very early phase (comparable to research on bibliometrics in the 1970s), the use of these data in research evaluation is already an issue. For example, altmetrics is considered in the Snowball Metrics project (Colledge, 2014). This project compiled a set of clearly defined indicators which will be used by participating universities (mostly Anglo-American universities) for research evaluation purposes. Also, many journals (e. g. Nature and PLoS journals) provide altmetric data for their papers on their webpages. Funders have also declared interest in using these metrics (Dinsmore et al., 2014). It seems that altmetrics will be used in practice before scientometrics research has produced standards on their reliable, fair and valid application (Weller, 2015).
This study uses one of the most important sources for altmetrics data, namely Mendeley. Mendeley “claims 3.1 million members. It was originally launched as software for managing and storing documents, but it encourages private and public social networking” (Van Noorden, 2014, p. 126). Since data from Mendeley can be received by an Application Programming Interface (API) without any problems and the coverage of the scientific literature has been pointed out as high (Priem, 2014), Mendeley is a very attractive data source for the reception of research. “Mendeley records the number of users that have listed it [i.e. an article], describing them as readers, whether or not they actually read it. Presumably, listing an article in Mendeley tends to reflect that an article has been read or will be read in the future, although there is no evidence that this assumption is true” (Thelwall & Maflahi, in press). Mohammadi & Thelwall (2014) found significant correlations between Mendeley reader counts and citation counts for the social sciences and humanities. Kraker et al. (2015) analyzed co-readership networks, and they briefly discussed the idea of bibliographic coupling and co-citation. In this study, we take a different approach as compared to Kraker et al. (2015), i.e. our focus is on readership coupling. Overviews about Mendeley readership studies have been presented by Haustein & Larivière (2014), Thelwall & Kousha (in press), and Thelwall & Maflahi (in press).
In order to produce the underlying data set, we match Mendeley data with data from F1000Prime. F1000Prime is a database with biomedical papers and their reviews by peers. It is intended as a support tool for researchers to receive hints for the most important literature. Since it is not clear who actually reads the F1000Prime recommended papers, we investigated the disciplines of researchers (and other people) who have read these papers. We are mainly interested in two questions: are F1000Prime papers only read by people from biomedicine or are people from other disciplines also interested? If so, which other disciplines show interest in F1000Prime papers? Which disciplines read F1000Prime papers frequently or seldom together? The latter question will be answered by using social network techniques.
F1000Prime is a post-publication peer review system of papers from medical and biological journals. This service is part of the Science Navigation Group, which publishes and develops information services for the professional biomedical community and the consumer market. Papers for F1000Prime are selected by a peer-nominated global "Faculty" of leading scientists and clinicians. The Faculty members rate the papers and explain their importance. This means that only a selected set of papers from the biomedical area covered is reviewed, and most of the papers are actually not (Kreiman & Maunsell, 2011; Wouters & Costas, 2012).
The Faculty nowadays numbers more than 5,000 members worldwide, assisted by further associates, which are organised into more than 40 subjects. Members can choose and evaluate any paper of interest; however, “the great majority pick papers published within the past month, including advance online papers, meaning that users can be made aware of important papers rapidly” (Wets et al., 2003, p. 254). Although many papers published in popular and high-profile journals (e.g. Nature, New England Journal of Medicine, Science) are evaluated, 85% of the papers selected come from specialised or less well-known journals (Wouters & Costas, 2012). The F1000Prime database is regarded as a useful aid for researchers (and other people working research-oriented) to obtain indications of the most relevant papers in the biomedical area: “The aim of Faculty of 1000 is not to provide an evaluation for all papers, as this would simply exacerbate the ‘noise’, but to take advantage of electronic developments to create the optimal human filter for effectively reducing the noise” (Wets et al., 2003, p. 253).
The F1000Prime publication set was provided to one of the authors in 2014. It consists of 149,227 records (papers and recommendations) with 114,582 unique papers which were published in various journals. 104,655 of those papers have a DOI and 112,983 of them have a PubMedID.
Within the first half of 2014 the reference manager Mendeley provided a new version of its API. Some restrictions of the previous API were lifted. For example, the usage statistics were previously provided in relative terms and only for the top three entries (Haustein & Larivière, 2014). The new API provides results in absolute numbers and not only for the top three but for all entries. Mendeley provides access to the readership status (e.g. professor, postdoc, or student) and the distribution of the Mendeley readership across scientific disciplines as well as countries via the API. Those sets of data can be correlated with other information available about papers (e.g. citations or Twitter counts).
Before one can start to use the Mendeley API, one has to register as a Mendeley user. Afterwards, registration of the desired application is necessary (http://dev.mendeley.com). Authentication with the API is done via OAuth 2.0. The credentials are set during registration of the application.
We used R (http://www.r-project.org/) to interface to the Mendeley API. It seems to us that using other interfaces does not change the functionality or responsiveness, but we did not try to use other interfaces. Mendeley provides sample codes for Javascript, Python, R, and Ruby (http://dev.mendeley.com/code/sample_code.html), whereby all requests to the API use HTTP GET and POST requests. Therefore, we suppose that any other scripting or programming language may be used. The reply is sent in Javascript Object Notation (JSON).
We requested user statistics for the F1000Prime publication set (n = 114,582 papers) using the PubMedID and DOI between the 4th and 6th of December 2014. Although the DOI (and possibly also the PubMedID) is not the unique identifier which it was intended to be (Franceschini et al., 2015), it is the currently best way to identify publications in the Mendeley API. If we could not found the PubMedID in the Mendeley database, the DOI was used. It is rather unlikely but possible that both identifiers are erroneous for the same paper. Therefore, we expect only an insignificant impact of erroneous DOIs and PubMedIDs on our results. We observed seemingly random connection problems. Sometimes those problems occurred after a few hundred or a few thousand requests. The largest chunk of requests we were able to get through the API without connection problems consisted of 47,629 papers. This large number of records is contrasted with smaller chunks of requests (between 1,049 and 9,307 records). Fortunately, data retrieval through the Mendeley API is very easy to restart. Therefore, we continued data retrieval with the same publication record where the connection problem occurred, so that no data loss occurred due to the connection problems.
Mendeley provides a breakdown of the user count into sub-disciplines. The possible values for disciplines and sub-disciplines can be obtained directly from the API via the GET /disciplines endpoint. Each discipline has a certain number of sub-disciplines. The sub-discipline “miscellaneous” occurs in every discipline. Each Mendeley user can select a discipline and a sub-discipline from a drop-down menu. This piece of information is not mandatory, like the user’s location.
Pajek is used to create the F1000Prime readership network (http://pajek.imfm.si/doku.php; de Nooy et al., 2011) applying the spring embedder of Kamada & Kawai (1989). Two reader counts from discipline A and at least two reader counts from discipline B constitute two links between both disciplines. From a matrix point of view, we have a symmetric readership coupling matrix, which has a two in row A and column B and vice versa for the aforementioned example. For detecting communities in the common readership of F1000Prime recommended papers, we used the VOS Clustering algorithm (Waltman et al., 2010), which is available in Pajek. The aim of this algorithm is to provide further insights into the structure of the network (Milojević, 2014). Our Pajek file is available free of charge at http://dx.doi.org/10.6084/m9.figshare.1386685.
It is not possible to distinguish if a Mendeley user only bookmarked a paper or also has read the bookmarked paper. However, for clarity reasons, we refer to bookmarks and observed reads as reader counts (following other studies). We found 6,263,913 Mendeley reader counts (on average 54.67 reader counts per paper) for the F1000Prime publication set. For 99.9% (n=6,257,603) of those reader counts, the discipline and sub-discipline information is also available. This is a much higher percentage than for the geographical location (Haunschild, et al., in press). For the F1000Prime publication set, the vast majority (74.94%) of Mendeley users is found in the “miscellaneous” sub-discipline of all disciplines. Therefore, we added up all the readers of all sub-disciplines for each discipline.
The results of our study are presented in Table 1. Nine disciplines contribute at least 1% to the reader counts of the F1000Prime publication set. The remaining 16 disciplines have less than 1%. As expected, most readers (81.78%) of the F1000Prime literature assign themselves to the biomedical (sub-) disciplines. All other disciplines comprise the remaining 15.19% of the F1000Prime readership at Mendeley. The third largest readership is found in the discipline psychology which is partly related to medicine. After chemistry, which is also related to biology, five other disciplines show readership values above 1% within the F1000Prime literature. Those disciplines seem rather unrelated to the field of biomedical research, especially environmental sciences (according to Figure 1). 3.05% of the F1000Prime reader counts (n = 190,919) at Mendeley come from other disciplines. The disciplines with most reader counts below 1% are: social sciences (0.67%), mathematics (0.42%), electrical and electronic engineering (0.27%), education (0.22%), and materials sciences (0.21%).
(sorted in decreasing order).
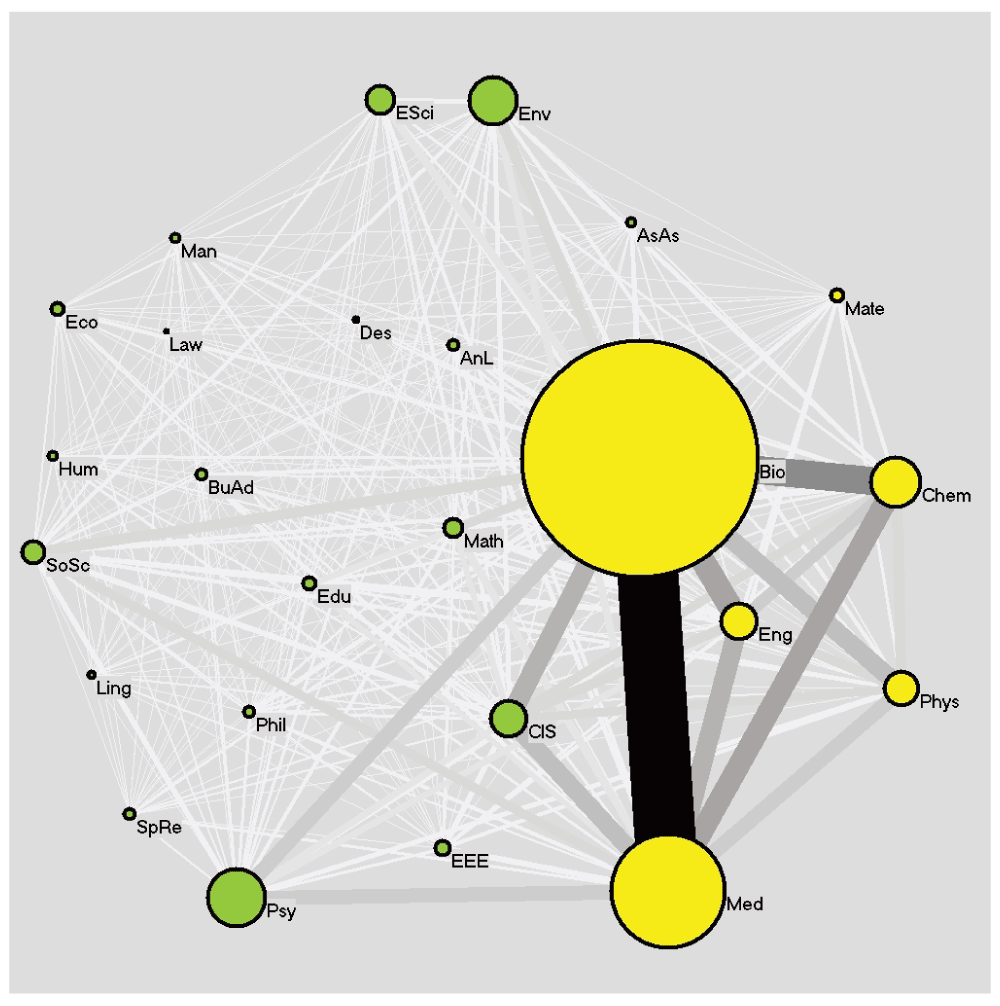
Network of F1000Prime recommended readers from arts and literature (AnL), astronomy and astrophysics (AsAs), biology (Bio), business administration (BuAd), chemistry (Chem), computer and information science (CIS), design (Des), earth sciences (ESci), economics (Eco), education (Edu), electrical and electronic engineering (EEE), engineering (Eng), environmental sciences (Env), humanities (Hum), law (Law), linguistics (Ling), management (Man), materials sciences (Mate), mathematics (Math), medicine (Med), philosophy (Phil), physics (Phys), psychology (Psy), social sciences (SoSc), sports and recreation (SpRe).
We also analyzed connections between the disciplines (see above). These are shown in Figure 1. A paper which is read by Mendeley users of different disciplines (e.g. biology and physics) constitutes a connection between these disciplines. Therefore, a paper which is read by Mendeley users of the same discipline does not contribute to the network system, but a paper which is read by Mendeley users of different disciplines contributes connections to the network. The size of the vertices in Figure 1 reflects the numbers of reader counts for each discipline. The thicker and darker the edges between two disciplines, the more frequently papers were saved by users of these two particular disciplines. One link is established between disciplines A and B if one reader count from both disciplines is observed for the same paper.
The location of the discipline vertex also informs about the connectivity. The closer the vertex is located towards the center, the more connections to different disciplines are found. There are 25 disciplines and 300 links among those disciplines in the dataset. With a density of 1, the network is rather dense. The average node degree is 24. According to Figure 1, the strongest connection shows up between biology (Bio) and medicine (Med). The disciplines computers and information science (CIS), engineering (Eng), and chemistry (Chem) have rather strong connections to biology (Bio) and medicine (Med). The discipline arts and literature (AnL) shows a low amount of readers (0.15%, close to sports and recreation with 0.16%) as well as a good connection to other disciplines in the network.
The community detection algorithm detected one dominant community in the network with biology, medicine, engineering, chemistry, and physics. Probably, the reader counts contributing to the green vertices in the network seem to be only weakly associated to the bio-medical literature. The disciplines shown as yellow vertices amount to 87.9% of the reader counts of the F1000Prime papers, while the remaining 12.1% reader counts are contributed by the disciplines shown as green vertices.
The (sub-) discipline of Mendeley readers is self-assigned and not mandatory. Still, we found that a large share (99.9%) of F1000Prime paper readers at Mendeley share their (sub-) discipline. Most readers (74.94%) assign the “miscellaneous” sub-discipline of their discipline to themselves. As the F1000Prime publication set is a collection of high-quality biomedical papers, we found – as expected – most readers in the disciplines of biology and medicine.
The network analyses revealed strong connections between engineering, chemistry, physics, biology, and medicine as well as their rather high reader percentages. These disciplines form a core set which can be differentiated from all other disciplines. In other words, besides this dominating set of disciplines no other set of strongly connecting disciplines could be identified. However, many of these other disciplines (e.g. mathematics, education, and arts and literature) are highly inter-connected (and connected with the core set), as their central location in the network indicates. Environmental sciences, psychology, and computer and information science are closely linked to biology although they are in a different community according to the employed algorithm. Chemistry and biology have a very strong link, much stronger than environmental sciences and biology. This is probably due to bio-chemical papers in the F1000Prime publication set.
Using a very specific data set, this study shows that Mendeley data can be used to investigate meaningfully the readership of a set of publications. Since the used data set here was from the biomedical area, the results agreed more or less to the formerly formulated expectations. Thus, it would be interesting in future research to use the new Mendeley API in order to investigate the readership of inter-disciplinary data sets or data sets for topics (e.g. climate change) which are inter-disciplinarily examined by researchers. Here, interesting insights can be expected using the new Mendeley API and the methods proposed in this study. For future studies, it would be also interesting to combine the discipline information from Mendeley with other user-specific data (the status group and the country of the users). This combination could lead to comparisons of discipline-specific networks between different countries (e.g. USA and India) and status groups (e.g. students and professors). However, for such comparisons the necessary data should be made available by Mendeley.
As expected, most Mendeley reader counts of F1000Prime publications can be associated with biology and medicine, although a significant percentage of reader counts originate from less related disciplines. According to the employed community algorithm 87.9% of the reader counts of the F1000Prime publications constitute the core of the readership of this publication set, while 12.1% of the reader counts are rather unexpected reader counts from less connected disciplines.
Figshare: Mendeley reader counts for F1000Prime papers and Pajek network file. Dois: 10.6084/m9.figshare.1301463 (Haunschild & Bornmann, 2014), 10.6084/m9.figshare.1386685 (Haunschild & Bornmann, 2015).
Wrote manuscript: RH and LB, Data acquisition: RH and LB, Data processing: RH, Data analysis: RH and LB, Produced graphics: LB, Manuscript revision: RH and LB.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 08 May 15 |
read | read |
|
Version 1 11 Feb 15 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)