Keywords
CENP-C, M18BP1, centromeric proteins, teleostei, kinetochore, CENP-C motif, cupin domain protein
CENP-C, M18BP1, centromeric proteins, teleostei, kinetochore, CENP-C motif, cupin domain protein
This update incorporates revisions suggested by the reviewer, includes common name categories of species names utilized in this study in the legend to figure 1 and makes a clear distinction between lobe-finned fishes in which CENP-C contains the "CENP-C motif" vs. non-lobe-finned fishes (specifically teleost fishes and shark) which lack an obvious "CENP-C motif".
See the author's detailed response to the review by Tatsuo Fukagawa
The kinetochore is a structure that connects chromosomal centromeric DNA to microtubules during mitosis and meiosis1. The centromere is epigenetically defined by the deposition of nucleosomes that contain the histone H3 variant CENP-A2. Centromeric protein CENP-C is required for both the recruitment of new CENP-A to the centromeric region as well as the initial assembly of the kinetochore. The CENP-C protein is generally considered to be ubiquitous in all eukaryotic taxa since homologs of CENP-C have been identified in yeast3 and Drosophila4 as well as many plants and vertebrates5. While CENP-C evolves so rapidly that very little homology is observed between distantly related taxa, a conserved CENP-C motif has been identified across all lineages studied5. This conserved motif should, therefore, be of utility to identify CENP-C orthologs in other species.
CENP-A has been initially shown to evolve adaptively in Drosophila6, in members of the Bressicaceae family7 and more recently in primates8 and in percid fishes9,10. CENP-C has also been shown to evolve adaptively in a number of animal and plant species5 as well as in primates8. In an effort to determine if CENP-C also evolves adaptively in fish species, searches were conducted in GenBank for Teleostei proteins that had been already identified as CENP-C or for genes that had been annotated as coding for CENP-C. No such teleost fish proteins or genes were found. BLASTP searches of just the Teleostei subset of GenBank were performed with the conserved vertebrate CENP-C motif and these too failed to find identified fish CENP-C proteins or genes. However, a set of orthologous C-terminal cupin domain containing genes have been identified in the elephant shark Callorhinchus milii and several teleost fish species that, while lacking most of the conserved CENP-C motif, have features that make these likely to be fish CENP-C orthologs.
Standard BLASTP searches were performed on the NCBI blast server. The vertebrate CENP-C motif NVRRTKRXRLKPLEYWRGERVBY used in BLASTP searches in this study was obtained from an alignment of 25 species including the lobe-finned fish Latimeria chalumnae, amphibians, reptiles, birds and mammals (Supplementary File S1). Retrieved sequences were aligned with the MUSCLE alignment feature in Geneious (version 6.1) sequence analysis software.
BLASTP searches with the vertebrate CENP-C motif identified CENP-C proteins from a variety of taxa, including plants, but did not identify any CENP-C in non-lobe-finned fish lineages. It is possible that CENP-C may be absent in ray-finned fish, but the ubiquity of this protein in other lineages and the central role of this protein in centromeric function make this unlikely. A C-terminal cupin domain protein encoded by a shark gene annotated in GenBank as CENP-C was used to identify homologs in Teleostei genomes by BLASTP. The retrieved teleost fish homologs were annotated as either calponin homology domain containing protein, neurofilament heavy polypeptide-like protein, or myb-like protein. The cupin domains of these proteins share significant homology to the cupin domain of other vertebrate CENP-C proteins (Figure 1). Within vertebrate CENP-C proteins the RxxRxxxxPLxYWxGERxxY sequence defines identities within the CENP-C motif located within about 100 amino acids upstream of the cupin domain (Figure 2). However, within the shark and teleost fish C-terminal cupin domain-containing protein sequences, only some of these CENP-C motif sequence identities were present (Figure 3) and, therefore, unambiguous identity of these proteins as CENP-C was not obvious.

Lobe-finned fish: Latimeria chalumnae. Amphibian: Xenopus laevis. Mammals: Homo sapiens, Equus caballus, Canis lupus familiaris. Reptile: Chelonia mydas. Bird: Gallus gallus. Shark: Callorhinchus milii. Teleost fish: Danio rerio, Oryzias latipes, Poecilia formosa, Poecilia reticulata, Maylandia zebra, Stegastes partitus.
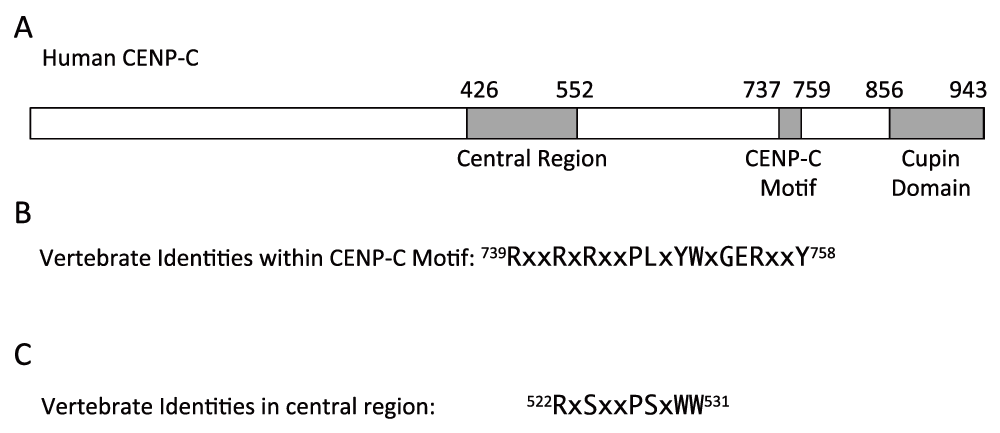
(A) Diagram of human CENP-C. (B) Amino acids that are identical in the CENP-C motif in vertebrates in which CENP-C has been identified. (C) Conserved sequence in the CENP-C central region that is homologous to part of the CENP-C motif. Amino acid locations within human CENP-C protein of conserved sequences are indicated at the beginning and end of each sequence.

(A) Diagram of putative Stegastes partitus CENP-C. (B) Alignment of fish central region sequences that contain the conserved RxxxxPxxWW sequence. (C) Alignment of the fish cupin domain proximal sequences that contain the conserved RxxxxPxxWW sequence. Amino acids matching the conserved sequence identities are highlighted in red. Amino acid locations within each species’ protein are indicated at the beginning and end of each sequence.
In a recent study that examined the interaction between CENP-C conserved domains and CENP-A containing nucleosomes (or nucleosomes containing histone H3 modified with a CENP-A C-terminal tail), Kato et al.11 identified within the conserved central region of CENP-C a RxSxxPSxWW consensus sequence (Figure 2) that is similar to the core portion of the CENP-C motif. Mutations of the arginine to alanine or the tryptophans to alanine in this sequence prevented the binding of this central region to the nucleosomes. So, functionally, the RxxxxPxxWW portion of the central region sequence is important to centromeric binding of CENP-C. Furthermore, mutations of the arginine, tyrosine and tryptophan in the core CENP-C motif RxxxxPxxYW also reduce the binding affinity the CENP-C to the nucleosomes11. A mutation of arginine to alanine in this core portion of the CENP-C motif was previously shown to prevent the binding of Xenopus CENP-C to centromeres12.
An alignment of the putative shark and teleost fish CENP-C proteins identified two conserved regions that contained the RxxxxPxxWW sequences (Figure 3). The placement of these sequences corresponds roughly to the locations of the central portion and the CENP-C motif of the vertebrate CENP-C (Figure 2). Therefore, it is likely that the combination of the C-terminal cupin domain and the presence of these centromeric nucleosome binding regions in positions generally corresponding to the locations of the central region and the CENP-C motif identifies these teleost genes as possible CENP-C orthologs. It will be necessary, of course, to verify if this protein is actually found at non-lobe-finned fish centromeres. It should be noted, however, that the distance between the cupin domain and the “CENP-C motif” position is about twice as long in the putative teleost fish CENP-C in comparison to this distance in CENP-C of other vertebrates. It is interesting that the putative shark “CENP-C motif” location lacks the tryptophans of the RxxxxPxxWW sequence and that Poecilia reticulata has a replacement of the first tryptophan in the conserved central region sequence (Figure 3). However, depending on other factors acting in the assembly of the centromere in various taxa, it may be possible that just one of those conserved RxxxxPxxWW sequences may be necessary for centromeric binding of the putative non-lobe-finned fish CENP-C. Indeed, the conserved central region is not present in chicken CENP-C13 and no homology to the RxxxxPxxWW portion of the conserved central region is detectable in CENP-C of other birds and reptiles. Yet a deletion mutant of chicken CENP-C in which the central portion had been removed was able to rescue CENP-C deficient chicken cells and also co-localized with CENP-T at centromeres13. This demonstrates that, at least in chickens, the central region is not necessary for CENP-A binding and, presumably, the C-terminal region which contains the CENP-C motif is sufficient for that purpose.
Interestingly, BLASTP searches of the Teleostei subset of GenBank retrieved centromeric protein M18BP1 sequences. This protein is recruited to centromeres by CENP-C14,15 and along with centromeric proteins Mis18α and Mis18β functions in the recruitment of CENP-A to centromeres16. The M18BP1 protein contains almost the entire vertebrate CENP-C motif in all vertebrates examined except in mammals (Figure 4). It appears that the CENP-C motif sequence is not exclusive to just CENP-C. Since both CENP-C and M18BP1 associate with centromeres and with each other, it is tempting to speculate that what has generally been regarded as a CENP-C motif sequence facilitates the interaction of both of these proteins with centromeric nucleosomes. Furthermore, since mammalian M18BP1 lacks this CENP-C motif, it is possible that mammalian M18BP1 may be more dependent on association with CENP-C to localize to the centromere than the M18BP1 of other vertebrate taxa.
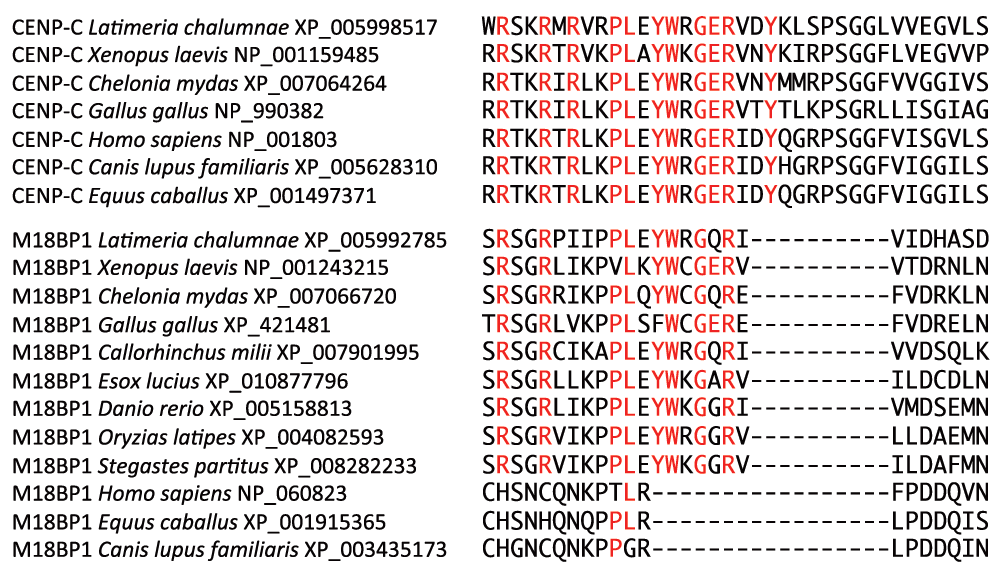
The vertebrate CENP-C motif sequence identities (Figure 2A) are highlighted in red.
This study was supported by a Faculty Research Grant funded by the University of West Georgia.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary File S1. Alignment of the C-terminal portion of vertebrate CENP-C proteins.
The vertebrate CENP-C motif containing consensus sequence utilized in BLASTP searches spans amino acids 11 to 33 and is highlighted in red.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
References
1. Carroll CW, Milks KJ, Straight AF: Dual recognition of CENP-A nucleosomes is required for centromere assembly.J Cell Biol. 2010; 189 (7): 1143-55 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 20 Jan 16 |
read | read | read |
|
Version 1 05 Aug 15 |
read | ||
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)