Keywords
latex, R, knitr, emacs, reproducible research, beta-hydroxybutyrate, melanoma, brain metastases
latex, R, knitr, emacs, reproducible research, beta-hydroxybutyrate, melanoma, brain metastases
One of the primary goals of any experimental research is to produce a nicely typeset document which explains the methods and results. This should be sufficient to allow the reader to recreate the work and thus to verify the results (given the correct tools).
In practice, much research is documented by adapting existing ‘office’-type software for this purpose (Microsoft, OpenOffice etc.). While there is much to be said for the ease of use of these techniques, they are not ideally suited to the purpose.
In particular, those that employ a ‘point-and-click’ graphical user interface (GUI) make it impossible to recreate these steps (mouse movements and clicks). The options for generating graphs and analyzing data are typically limited and often require the use of separate ‘third-party’ software for these steps (e.g. SPSS, GraphPad Prism). This again makes the reproduction of results a challenge.
There are many free and open source alternatives which are designed with the needs of the laboratory researcher in mind. Ease of use appears to be the principle reason for their lack of widespread use. The mp (for ‘make-pdf’) package came about as an attempt to bring elements from a number of these diverse sources together under one roof. It was also motivated by the repetitive nature of much laboratory research. Successive experiments often differ little in method, so that the analysis often uses the same techniques, with new experimental data being supplied each time.
Details of the experiment are documented and the R code for analysis is checked as usual; the mp package automates much of the rest of the process with one keystroke. Typesetting is performed using LaTeX, which has become the industry standard for publishers of scientific journals1.
Emacs is the text editor which brings these methods together. Emacs itself has been criticized for lack of ease of use, although if used purely as a text/file editor, as in the examples here, it remains quite simple2. Some familiarity with setting customizable variables is required when moving beyond the default settings for the mp package.
The examples below do assume some familiarity with R. The transition from familiar GUI-style data-analysis to terminal-based output may appear daunting at first. For those considering taking the plunge, we hope that these simple examples will help to illustrate how easy the R language can be to use. As a long-term investment, we feel that the time taken to become familiar with these methods is likely to be more than compensated by subsequent improvements in the speed and simplicity of workflow.
There are already a number of tools useful in converting R source code to a nicely displayed .pdf file. The goal of the mp package is to generate such .pdfs ‘at the touch of a button’. It allows for conversion of the source .R file to the final .pdf3. Further editing, when required, is performed on an intermediary (or ‘go-between’) file - either a no-web (.Rnw) or an .org file4,5. mp can also generate documentation for an elisp package from the package’s own source code.
Currently, there are three widely used converters when generating a .pdf from an .R file. These are:
Following the existing trend to name these tools after processes involved in fabric making, we refer to them collectively as ‘entwiners’.
An allied approach, converting from .org to .Rnw has also been implemented using the ravel package for Emacs6. Another similarly themed project is the pander package for R, which integrates R with the Haskell library pandoc7,8. This could serve as an alternative to any of the above or be used in conjunction with them. For the sake of simplicity, the range of alternatives provided by the following three ‘entwiners’ appears sufficient for most needs.
Sweave. The oldest and best-supported of these converters is Sweave9. It is arguably the best integrated with R and remains the standard tool for R package developers writing accompanying vignettes. It remains widely used in generating statistical reports.
It does suffer from a number of limitations relative to its counterparts. In particular, the displayed code ‘as-is’ has little formatting or use of color.
Also, by default, only one figure per ‘chunk’ (i.e. block of code) is supported. Outputting multiple figures typically means writing to, and reading from files explicitly in R, which can be tedious to implement.
Knitr. The knitr package for R generates code for display which is arguably ‘prettier’ than Sweave10. It also allows for multiple figures per chunk. It also allows the chunks to be kept in a separate source file - as opposed to requiring them to be part of the .Rnw file. There are more options for the display of terminal output e.g. including error messages. Like Sweave, it can be used to build R package vignettes.
knitr already integrates well with some existing GUI-style .tex editors, particularly LyX. The latter is part of ‘Scientific Workplace’ (SW), which, like mp, tries to make life easier for the laboratory researcher by providing a simplified workspace. In the case of SW, a GUI is preferred to directly editing files11.
org-mode. .org files are arguably easier/more intuitive to read and edit than .tex, particularly for users new to the latter. In particular, tables are much more straightforward to read, create and modify. Also, the use of ‘collapsible’ section headers makes it easy to see the structure of a document at-a-glance before focusing on one section for editing.
Org-mode in Emacs allows for the export of code chunks/blocks. Like Sweave, org-mode chunks suffer from the drawback that multiple figures per chunk are not supported by default.
While org-mode loses the attractive code printing of knitr, some worthy alternatives are provided by the LaTeX package listings12. These include the option to include LaTeX maths markup in the code commentary, for example to display equations.
Org-mode allows for conversion/export to multiple file types. By default, mp converts .tex to .pdf but alternatives are straightforward, such as to .html or to .MARKDOWN.
In mp, the settings adopted for listings are modeled on those of the knitr defaults. We acknowledge that they are perhaps not as attractive. These are the same settings used for the code listings in this article.
An alternative to listings is the minted package for LaTeX. While probably more attractive for code display in general, its use involves a package for another language (Pygments, written for python). Also, outlining multi-page blocks with background coloring is tricky to automate.
Much scientific research is reported in the form of a summary and it is not traditional for researchers to provide detailed accounts of the original experiments along with the original observations/results made at the time.
The classic quote on the subject is from Buckheit and Donoho13:
An article about computational science in a scientific publication is not the scholarship itself, it is merely advertising of the scholarship. The actual scholarship is the complete software development environment and complete set of instructions which generated the figures.
The same may well be said of biology. With a traditional print-form journal, the constraints of space limit the length of articles and thus providing content as a summary is sensible. However, the option to refer to online-only Supplementary material now obviates such impediments.
Such supplements may be be valuable for those seeking to reproduce steps in the research. They may also may be of interest to researchers working on closely related problems, particularly by saving time on preparatory work. Providing original data may allow such data to be combined with that from subsequent experiments. Also, analytic techniques beyond those used in the original paper may later be brought to bear by other researchers.
The details of individual experiments, particularly ‘pilot’ studies of an exploratory nature or those which yielded negative results are likely to be of limited interest to a general readership. Including these resources as supplements appears reasonable.
The supplements of this main paper illustrate the applicability of the mp software to a series of experiments from our laboratory which demonstrate the inhibitory effects of beta-hydroxybutyrate (BHB) on the growth and migration of a melanoma cell line, selected for its propensity to form brain metastases, in vitro.
Package details. mp is written in elisp, the native language of Emacs14. The package aims to automate the workflow associated with converting from .R files to .pdfs. Data analysis is done primarily from the .R file directly - in preference to the intermediary .Rnw or .org file, which can become cumbersome.
mp is ‘opinionated’ when generating intermediary files. Certain defaults have been chosen based on the authors’ experience, for example with the settings for the LaTeX hyperref package.
In order to streamline the whole process, most of the processing takes place asynchronously. That is, document editing can proceed as the Emacs terminal is not ‘frozen’ during the process of .pdf generation.
No support for caching is provided, although with short documents similar in scale to the examples below this should not result in much loss of performance. The time to compile is <10 secs with an Intel i5-2430M processor for all of the examples given.
The manual for the mp package is given as Supplementary material (this is generated from the code itself, using an .org intermediary file.).
The package defines the mp minor mode for Emacs. The only keybinding involves the function mp-mp with Ctrl-Alt-|(usually found above the ‘Return’ key; in Emacs parlance this is also known as C-M-|). This is intended as a gateway to all of the package functions, all of which can also be run individually (or ‘interactively’).
mp-mp will prompt for a filename; if none is supplied, it will look first at the current buffer. If this is not an .R, .Rnw, .org.tex or .el file it will select the appropriate file from the default-directory as that which has most recently modified. Thereafter it will search up the directory tree if no such file is found.
Instead of a filename, the single character ‘p’ may be given to display the appropriate .pdf associated with the current file or directory.
Control flow. A simplified flow diagram of the mp package is presented in Figure 1.
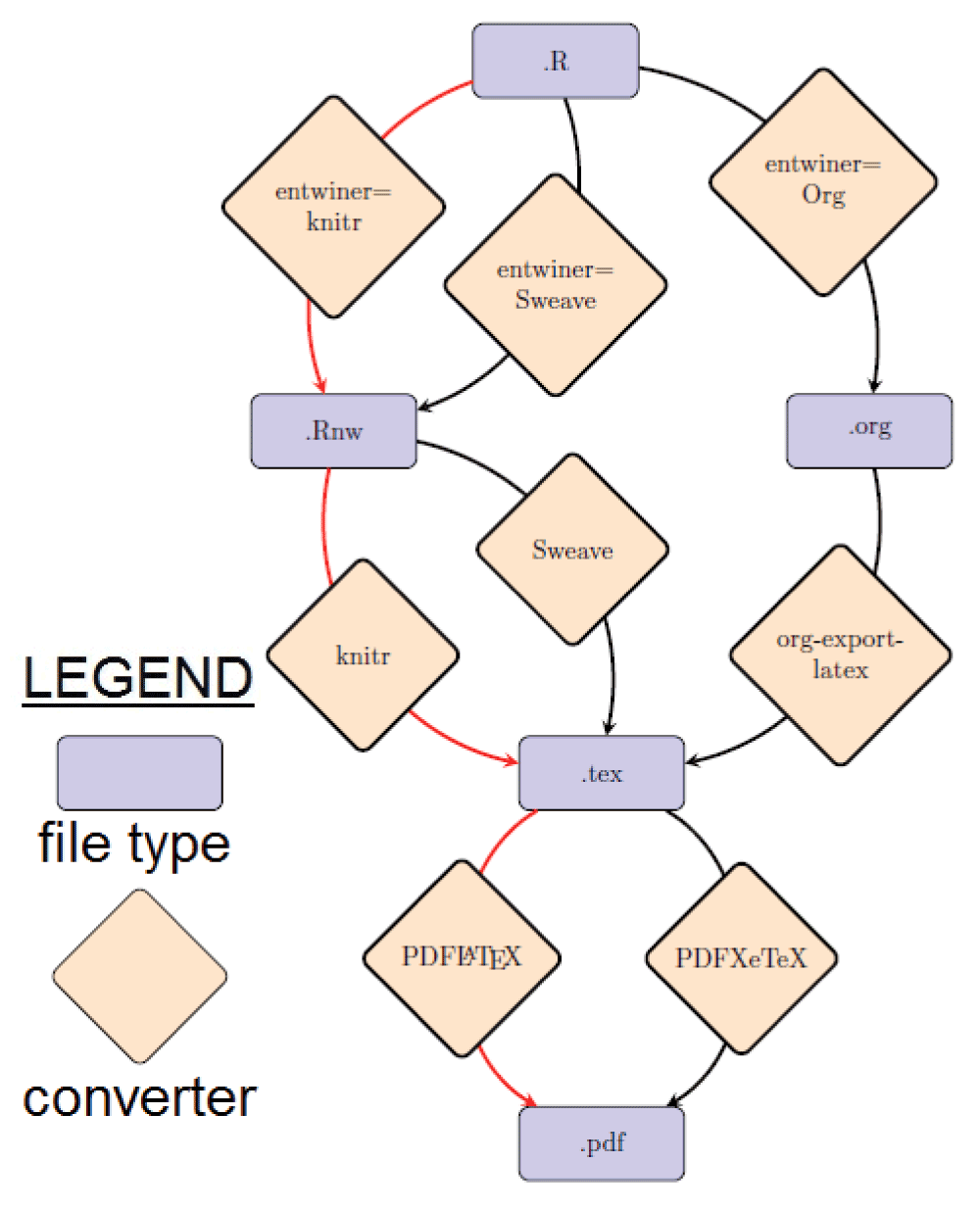
The red arrow shows the default flow path.
A more detailed diagram, which also shows the customizable variables, is available as Supplementary material (mpFlow.pdf). While the latter may appear complex, all of the functions and variables are accessed through the ‘umbrella’ mp-mp function so that in a typical use case there is no need to alter the default settings.
For more experienced users, this use of multiple functions and variables allows for highly granular control, if desired.
Use case. A typical use-case for experimental work proceeds as follows:
1. Create a new directory to hold all the files below.
2. Draft the code for analysis in .R. Arbitrary ‘placeholder’ data may be used at this stage.
3. Create a ‘skeleton‘
4. Use the intermediary file generated to draft the experimental protocol.
5. Complete the experiment.
6. Update the .R or intermediary file generated (.Rnw or .org) with the results of the experiment and the conclusions.
7. Generate the final .pdf from the intermediary file.
Typically, the user will begin with an .R file which contains a number of chunks of R code. These are designated by the header ## ---- chunkName. This follows the convention introduced by knitr for naming chunks. Each chunk continues until the following chunk starts.
The chunks are processed by the option given by the customizable variable mp-entwiner, which is knitr by default.
If no .Rnw file exists in the current directory, one is created by mp-skeleton-nw. If an .Rnw file does exist, it is updated using mp-update-nw; those chunks matching the names in the .R file are updated and new chunks are added if required.
The default workflow is shown in Figure 1 by the path through the red arrows. Some of the pros and cons of different choices of mp-entwiner are shown in 11.
‘Hello world’ with mp. We begin with the simplest use case. We create the directory hello and place the contents of Listing 1 into hello.R. With this file open in Emacs, we then run mp-mp with the key combination C-M-|.
Listing 1: The file hello.R## ---- Print hello print(”Hello world”) ## ---- Here are two graphs ## here’s a comment plot(1:10, 10:100) ## this second graph should appear ## below the first plot(1:10, sqrt(1:10))
The final product, hello.pdf is shown in Figure 2.
The intermediary file hello.Rnw will be left open in Emacs for further modification and this is shown in Listing 2. There is a lengthy preamble which sets defaults for multiple options, here in LaTeX and R (including knitr). Of course, most of this is not relevant to such a simple example.
Listing 2: The file hello.Rnw% \documentclass{article} % % modified from default setup for knitr % \usepackage[]{graphicx} \usepackage[]{color} \usepackage{framed} % recommended with ‘knitr’ \usepackage{alltt} \usepackage{mathtools} \usepackage[sc]{mathpazo} \usepackage{geometry} \geometry{verbose, tmargin=2.5cm, bmargin =2.5cm, lmargin=2.5cm, rmargin=2.5cm} \setcounter{secnumdepth}{2} \setcounter{tocdepth}{2} \usepackage{url} \usepackage{hyperref} \hypersetup{unicode=true, pdfusetitle} \hypersetup{bookmarks=true, bookmarksnumbered=true} \hypersetup{bookmarksopen=true, bookmarksopenlevel=2} \hypersetup{breaklinks=false, pdfborder={0 0 1}} \hypersetup{backref=false} \hypersetup{colorlinks=true} \definecolor{myDarkBlue}{rgb}{0, 0, 0.5} \hypersetup{linkcolor=myDarkBlue} \hypersetup{pdfstartview={XYZ null null 1}} % % Palatino family \usepackage[T1]{fontenc} \usepackage{mathpazo} \linespread{1.05} \usepackage[scaled]{helvet} % % other useful additions % %% for rerunfilecheck: % no need to rerun to get outlines right \usepackage{bookmark} %% or nice tables \usepackage{booktabs} %% for e.g. \formatdate \usepackage{datetime} %% for SI units \usepackage{siunitx} %% for chemical symbols \usepackage[version=3]{mhchem} %% to use forced ’here’ %% e.g. \begin{figure}[H] \usepackage{float} %% for large numbers of floats \usepackage{morefloats} %% to keep floats in same section \usepackage[section]{placeins} %% for tables > 1 page \usepackage{longtable} % %%---------------------------------------- % \begin{document} % % knitr chunks <<setup, include=FALSE>>= library(knitr) ### Set global chunk options opts_chunk$set( eval=TRUE, ## text results echo=TRUE, results=c(’markup’, ’asis’, ’hold’, ’hide’) [1], collapse=FALSE, warning=TRUE, message=TRUE, error=TRUE, split=FALSE, include=TRUE, strip.white=TRUE, ## code decoration tidy=FALSE, prompt=FALSE, comment=’##’, highlight=TRUE, size=’normalsize’, background=c(’#F7F7F7’, colors()[479], c (0.1, 0.2, 0.3))[1], ## cache cache=FALSE, ## plots fig.path=c(’figure’, ’figure/minimal-’), fig.keep=c(’high’, ’none’, ’all’, ’first’, ’last’)[1], fig.align=c(’center’, ’left’, ’right’, ’default’)[1], fig.show=c(’hold’, ’asis’, ’animate’, ’hide’)[1], dev=c(’pdf’, ’png’, ’tikz’)[1], fig.width=7, fig.height=7, #inches fig.env=c(’figure’, ’marginfigure’)[1], fig.pos=c(’’, ’h’, ’t’, ’b’, ’p’, ’H’)[1]) ### Set R options options(formatR.arrow=TRUE, width=60) knit_hooks$set(inline = function(x) { ## if (is.numeric(x)) return(knitr:::format _sci(x, ’latex’)) highr::hi_latex(x) }) ## uncomment below to change theme ## knit_theme$get() ## opts_knit$set(out.format=’latex’) ## thm1 <- knit_theme$get(’acid’) ## knit_theme$set(thm1) @ % knitr read chunks <<readChunks, include=FALSE>>= read_chunk(’hello.R’) @ \title{hello} \author{Chris Dardis} \maketitle % page numbers appear top-right \pagestyle{headings} \tableofcontents \subsection{Print hello} <<Print hello>>= @ \subsection{Here are two graphs} <<Here are two graphs>>= @ % %% \bibliographystyle{plain} %% \bibliography{hello} \end{document}
Examples of use. Some examples more relevant to the laboratory follow. These are summarized in Table 2. These are all available as Supplementary material and we encourage the reader to refer to the relevant files while reading the present article. Further details of the experimental work are given at the end of this section.
| File | Entwiner | Edit | TeX | Final | |
|---|---|---|---|---|---|
| 1 | 8k | knitr | .Rnw* | LaTeX | .pdf* |
| 2 | 16k | knitr | .Rnw* | LaTeX | .pdf* |
| 3 | 16k2 | Sweave | .Rnw* | XeTeX | .pdf* |
| 4 | 32k | Org | .org* | LaTeX | .pdf* |
| 5 | 32k2 | Org | .org* | LaTeX | .html* |
| 6 | sa | knitr | .Rnw* | XeTeX | .pdf* |
* = file available as Supplementary material
The ‘file-stems’ (i.e. the filename without a suffix) refer to the starting quantity of cells per 12-well plate. ‘sa’ in the last example stands for ‘scratch assay’.
The variable mp-preamble is used to set some defaults useful for typesetting documentation for the laboratory. For example, the LaTeX package siunitx is used for the correct display of scientific units15.
Example 1: 8k.pdf
Experiment: Establish number of cells required to establish sustained growth. Start with 8000 cells per 12-well plate. Our first aim was to establish the optimal number of B16-F1-Luc2-BR2 cells with which to seed a growth curve. We wished to establish sustained growth over the period of the experiment, without reaching a ceiling (i.e. maximum density).
Typesetting: default settings. The file 8k.R is given in Listing 3:
Listing 3: The file 8k.R## ---- data (df1 <- data.frame(Day = rep(0:4, each=3), Well = rep(LETTERS[1:3], 5), Count = c (rep (8e3, 3), 193e3, 148e3, 159e3, 78e3, 63e3, 79e3, 33e3, 56e3, 33e3, 13e3, 15e3, 10e3))) ## ---- stdError stdErr <- function(x) sqrt(var(x)) / sqrt( length(x)) library(plyr) (df2 <- ddply(df1, .(Day), function(df) c(mean = mean(df$Count), SE = stdErr(df$Count), ’mean+SE’ = mean(df$Count) + stdErr(df$Count), ’mean−SE’ = mean(df$Count) - stdErr(df$Count)))) ## ---- plot library(plotrix) # confidence interval plot with(df2, plotCI(x=Day, y=mean, ui=mean+SE, li =mean-SE, xlab=”Day”, ylab=”Cell counts (Cells/ ml)”, axes=FALSE)) axis(side=1, at=c(0:4)) axis(side=2, at=seq(0, 150e3, by=50e3)) box() with(df2, lines(Day, mean))
With the above file open, we call mp-mp to generate the a.pdf; page 1 of the result is shown in part in Figure 3.
As an intermediary step, the file 8k.Rnw is generated. This is an R no-web file, which is essentially a typical .tex file with the additional code chunks inserted4. The term ‘web’ is not in reference to the world-wide web but to distinguish it from a contemporaneous approach to literate programming known as ‘web’.
We modify this to add details of the experimental protocol, written in LaTeX. The principal modifications are the use of 12pt for typesetting, which we adopt from hereon and the inclusion of some graphics with the LaTeX subfig package16.
With 8k.Rnw still open, we call mp-mp again to generate the final .pdf.
Example 2: 16k.pdf
Experiment: Number of cells required to establish sustained growth. 16000 cells per 12-well plate. Again, we tried to establish the optimal number of B16-F1-Luc2-BR2 cells with which to seed a growth curve. We increased the starting number of cells per well from 8000 to 16000. This appeared to be more promising.
Typesetting: using R to generate .tex output. We again begin with an .R file. This time however, we use R to create LaTeX output, rather than displaying the results of a chunk as R code. The output from R is interpreted as LaTeX directly - rather than as typical terminal output. We use the R xtable package for this purpose; there are many good alternatives17.
An example of R code generating output which can be interpreted by LaTeX is shown in Listing 4.
Listing 4: From the file 16k.R## ---- Cell counts on Day 0 df1 <- data.frame(Cells_per_ml = c(392e3, 332e3, 300e3)) library(xtable) print(xtable(df1, caption=”Cell counts on day 0”, align=c(”l”, ”c”), display=c(”d”, ”e”), label=”tab:ccd0”), booktabs=TRUE)
Again, the .Rnw file needs some modification. The main change is to set the knitr option results= ‘markup ’ to results= ‘asis ’ to allow the results to be interpreted as .tex.
We can also restructure the document to allow the code output (here, in the form of tables or single lines of terminal output) to be mixed with the main text. This ensures there is no needless duplication of data entry, as is the case in the earlier example.
The output is shown as Supplementary material in 16k.pdf. Most of the materials and methods are unchanged from 8k.pdf.
Example 3: 16k2.pdf
Experiment: Demonstrate impaired growth in the presence of BHB. 16000 cells per 12-well plate. We sought to determine whether the growth of B16-F1-Luc2-BR2 cells would be impaired in the presence of BHB at a concentration of 10 mmol/L.
Due to the large standard error on day 4, no difference could be demonstrated via a t-test. However comparing linear models with and without treatment did show a significant effect (analysis of variance (ANOVA) p = 0.048).
Typesetting: Using Sweave and XeTeX. Here, we change the variable mp-entwiner to Sweave. We also change mp-latex to XeLaTeX to allow the use of an OpenType font (by default this is ‘Linx Libertine’). While certain OpenType fonts are available as packages for LaTeX, XeTeX allows for a greater range and greater flexibility.
As in example 2, the output from each chunk is again typically interpreted as .tex (rather than displayed as R code) by including results=tex in mp-Sweave-opts.
Some modifications to the .Rnw file generated follow. There is one chunk where we prefer the output to retain typical R code formatting. Also we need to change the variable mp-Sweave-opts to fig=TRUE for the chunk which produces a plot.
This example also shows the value of LaTeX for typesetting equations - calculation of correct masses are nicely displayed and easy to follow. This is shown in Figure 4.
Example 4: 32k.pdf
Experiment: Demonstrate impaired growth in the presence of BHB. 32000 cells per 12-well plate. As the preceding experiment was inconclusive, we tried the same technique again, this time using a higher number of cells to start with.
Unfortunately, no observations were taken on day 3 of the experiment. This meant that both the t-test on day 4 and the ANOVA comparing linear models were not significant, although the latter did come close (p = 0.07).
Typesetting: using Org. Changing mp-entwiner to Org allows us to use an .org file as the intermediary step.
The output from this process is given as 32k.pdf. Again, some minor modifications to the intermediary file are required. In this case we need to change the headers for the chunk which produces a plot to specify the output of graphics (as opposed to text) and to specify an external file for the graphical output.
We also change some of the results to latex and pp (pretty print) to give examples of different styles of output. 32k.pdf also features a printout of help for an R function.
Example 5: 32k2.pdf
Experiment: Demonstrate impaired growth in the presence of BHB. 32000 cells per 12-well plate. We repeated the preceding experiment and this time there was a conclusive difference between the cell counts in those grown with BHB vs. controls.
Typesetting: Using Org to generate HTML output. This example shows how the .org intermediary can be converted to .html as an alternative to .pdf. Converting to HTML means we lose all but the simplest LaTeX commands and so the document needs to be written in a simpler style. While we lose some of the nice typesetting features of LaTeX, this simplicity has its own attraction.
These files are faster to produce and smaller than their .pdf counterparts (1/5 the size in this example). HTML is also likely to be simpler to integrate into an existing website and offers the possibility of almost ‘real-time’ reporting of experimental results.
Example 6: sa.pdf
Experiment: Demonstrate impaired cell mobility in the presence of BHB. Here we use a scratch assay18. A line is made in the center of the wells on a 12-well plate with confluent cell growth. Images taken at various timepoints are analyzed with freely available imageJ software. The distance between the cells on either side of the scratch decreases as they migrate back to the center. This corresponds to a decrease in ‘image density’ as measured by imageJ. This may be downloaded from:
http://rsbweb.nih.gov/ij/download.html
Typesetting: Emulating a laboratory notebook with XeTeX. The final example shows how a cursive font may be used to mimic a handwritten lab notebook. We set mp-entwiner to knitr and set mp-latex to XeTeX to allow the use of an unusual font. A sample is shown in Figure 5.
When combining multiple images on a single page, fitting them into the cells of a table can be helpful for clarity and this example features a number of such tables.
System requirements: this should work with any recent version of Emacs, which is platform independent (i.e. works on Windows, Linux, Mac-OS). Version ≥ 24.4 is recommended to allow for automated export of .org to .tex.
To export to HTML, the elisp package htmlize is required19. Links to the package source may be found at:
http://emacswiki.org/emacs/Htmlize
A recent installation of R (>3.0) and TeX (2013 and on) is also assumed. We used TeX Live 2013 for these examples.
The package should work with LuaTeX as well as LaTeX, although this has not been thoroughly tested.
The purpose of these experiments was to demonstrate an inhibitory effect of BHB on the growth and migration of a melanoma cell line in vitro. Details of the aims, methods, results and conclusions are given for each experiment separately (see Table 2).
A summary of these experiments would normally make up just one part of an article; further details of the individual experiments could be combined and made available as a supplement. We provide the experiments separately to provide a typical use-case for the mp package.
Cells. B-16 melanoma cells were obtained from American Type Culture Collection (ATCC). To facilitate quantitative measurement of tumor growth, they were modified as described previously20. Briefly, the cells were stably transfected with the gene encoding luc2 (luciferase) using the pGL4.51 [luc2/CMV/Neo] vector (Promega Corp, Madison, WI) and FuGENE 6 Transfection Reagent (Roche Applied Science, Indianapolis, IN) following conditions specified by the manufacturer. They were then injected into the heart (the right ventricle) of a mouse. These animals were sacrificed when bioluminescence was detected in the brain. Cells metastatic to the brain were recovered and were put into culture.
The same procedure was repeated again (injection into mouse heart and recovery of brain metastases). These cells are designated B16-F1-Luc2-BR2.
Animal handling. This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of St. Josephs Hospital and Medical Center (protocol number 334 (A3510-01)). All surgery was performed under ketamine/xylazine anesthesia, and every effort was made to minimize suffering.
Effects of BHB on growth. Firstly, it was necessary to determine the optimal starting number of cells to use for a growth curve. If the cells are plated too sparsely, they have a tendency not to grow. Likewise if the concentration is too great to start with, then this will make any difference due to the effect of BHB difficult to see. This is complicated further by growing the cells in a circular well - the cells are in closer communication close to the center and more separated close to the edge. In choosing a starting number, we were guided primarily by prior work in our own laboratory.
To determine whether BHB would inhibit cell growth, we chose a concentration of 10 mmol/L, again based primarily on our prior experience. Under normal conditions, serum BHB has an upper reference limit of 0.3–0.5 mmol/L. Tests of BHB concentration (for patients) are available commercially and can be used in by those adhering to the ketogenic diet. This diet is most commonly employed in the treatment of drug-resistant epilepsy, where serum BHB concentration has been shown to correlate with the degree of seizure control21. In this study of 74 children, median levels were 8–10 mmol/L.
We chose a concentration of 10 mmol/L in order to be certain that an effect was present, before considering whether lower levels would be similarly efficacious.
Effects of BHB on migration. The scratch assay is a simple and widely used test to assess cell migration. Details of a typical protocol for the technique are available18. Using the same cell line, we sought to illustrate impaired migration in the presence of BHB.
As shown in Figure 6, we were able to demonstrate a difference in growth rates with an initial concentration of cells of 32,000 per well and a BHB concentration of 10 mmol/L. This is shown in full in 32k2.html.
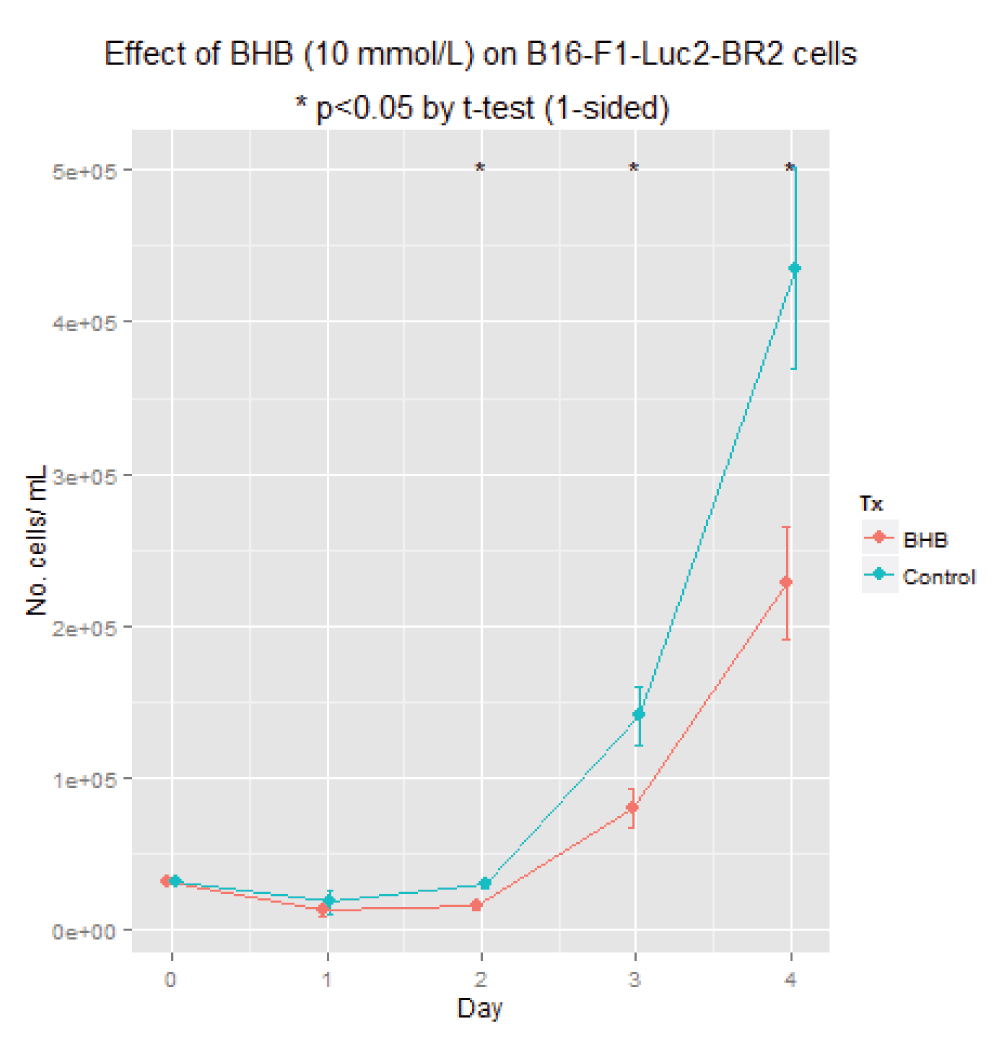
Taken from 32k2.html (Supplementary material).
We were also able to demonstrate a decrease in cell migration using the scratch assay, as shown in Figure 7, which is taken from sa.pdf.
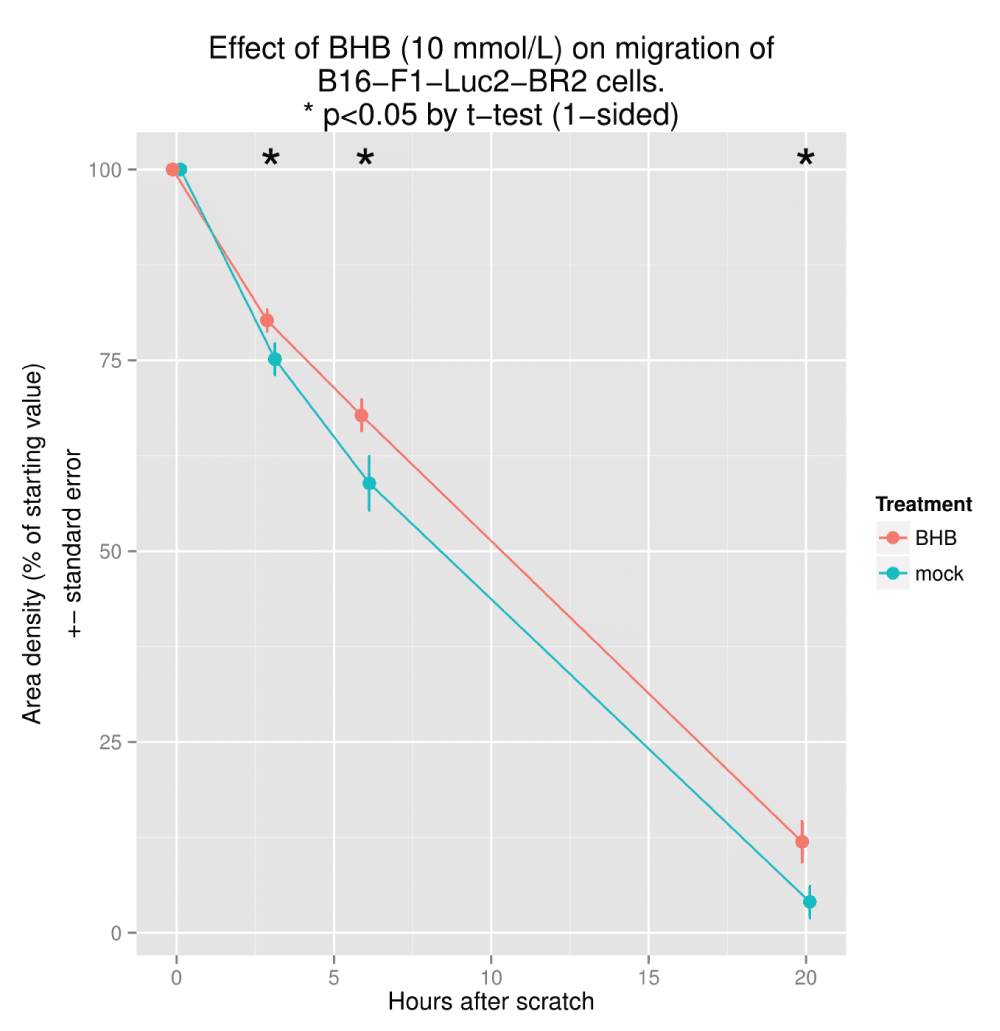
Taken from sa.pdf (Supplementary material).
The inhibitory effects of BHB on the growth of neoplastic cells has now been observed in a number of cells lines in our laboratory and those of other investigators22. Similarly it has been shown to impair apoptosis and necrosis of the L929 mouse cell line (taken from normal subcutaneous areolar and adipose tissue)23.
Impairment of growth of neoplastic cells due to ketones has been suggested to be in part due to the polyacetylation of histones24. Ketone bodies also impair glycolysis, on which neoplastic cells are more heavily dependent. This occurs in part through inhibition of the activity of phosphofructokinase and hexokinase25,26.
The finding that inhibition of growth occurs in a cell line selected for their propensity to metastasize to the brain is novel. Brain metastases are a significant contributor to morbidity and mortality in patients with cancer27. We feel that this approach deserves further investigation. In particular, it would be of interest to determine whether the same phenomenon would be observed in breast and lung cancer cell lines with a propensity to metastasize to the brain. It would also be interesting to determine whether the inhibitory effects of BHB on cancer cells are evident in vivo when ketones are given as a dietary supplement with a relatively normal diet - as opposed to the ketogenic diet, which may be challenging to adopt for patients affected by cancer.
We have demonstrated here that BHB, at a concentration of 10 mmol/L, is inhibitory to the growth and migration of B16-F1-Luc2-BR2 melanoma cells that were selected for their propensity to form brain metastases.
The mp package appears to be a useful addition to the tools available for facilitating reproducible research. Future development will likely include increased support for the LaTeX package biblatex. Collaboration and suggestions for improvement are welcome. Please try to address these to the development site on GitHub if possible.
The datasets for all experiments are available as part of the Supplementary material.
One of the primary motivations for developing this package was to facilitate sharing of datasets such as these.
CD - carried out the experiments (except for the scratch assay), wrote the software, performed the data analysis and drafted the manuscript.
ECW - carried out the scratch assay experiment; he was also closely involved in the design and execution of the other protocols.
ACS - supervised the experimental protocols and interpreted the results.
All authors have revised and approved the final manuscript.
The authors would like to thank Students Supporting Brain Tumor Research for their support.
Manual for mp package. Documentation, including all functions and variables in the package. Generated from the source code. This package is ‘self-documenting’. mp.pdf is the manual for the package, which was generated with mp-el-tex. The option INCLUDESOURCE is set to t, so that the source code is shown for all functions. The intermediary file is .org which allows package documentation to be nicely displayed in list or table format using TeX, as appropriate.
Click here to access the data.
Flow diagram for mp package. An image, showing the control flow for the package, including file types, functions and customizable variables.
Click here to access the data.
Establish a 5-day growth curve starting with 8,000 cells/well. Example 1; default settings. Intermediary .Rnw file, typeset with knitr.
Click here to access the data.
http://dx.doi.org/10.5256/f1000research.6800.s97727
Establish a 5-day growth curve starting with 16,000 cells/well. Example 2; using R to generate LaTeX output. Intermediary .Rnw file, typeset with knitr.
Click here to access the data.
http://dx.doi.org/10.5256/f1000research.6800.s97728
Effects of BHB on growth starting with 16,000 cells/well. Example 3; using Sweave. Intermediary .Rnw file, typeset with Sweave (and XeTeX).
Click here to access the data.
http://dx.doi.org/10.5256/f1000research.6800.s97729
Effects of BHB on growth starting with 32,000 cells/well. Example 4; using org-mode. Intermediary .org file, typeset with org-mode and listings.
Click here to access the data.
http://dx.doi.org/10.5256/f1000research.6800.s97730
Effects of BHB on growth starting with 32,000 cells/well. Example 5; using org-mode to generate .html output. Intermediary .org file, typeset with org-mode and listings.
Click here to access the data.
http://dx.doi.org/10.5256/f1000research.6800.s97731
Effects of BHB on migration, via scratch assay. Example 6; using a cursive font and placing images in a table. Intermediary.Rnw file, typeset with knitr (and XeTeX).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 30 Mar 16 |
read | |
|
Version 1 05 Aug 15 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)