Keywords
Breast cancer, NKILA, PMEPA1, NF-ĸB, long noncoding RNA, antisense, promoter, evolution
Breast cancer, NKILA, PMEPA1, NF-ĸB, long noncoding RNA, antisense, promoter, evolution
Liu et al.1 investigated breast cancer cell lines for possible association of known long noncoding transcripts with immunostimulation. They found that an unspliced lncRNA, which they designated “NKILA” (represented by large yellow box in Figure 1), could be upregulated by several immune agents. However, they did not mention the existence of a reported spliced form of NKILA (GenBank accession DA866558, see Figure 1), or, - our major criticism - that NKILA is divergently transcribed from prostate transmembrane protein, androgen induced 1 (PMEPA1) and antisense to some of its transcripts (see Figure 1). One of the alternative names for PMEPA1 is solid tumor associated gene 1 (STAG1)2, and its expression was found upregulated in several tumors including breast cancer (e.g., references 2 and 3). Liu et al.1 found that the NKILA promoter region contains an NF-κB binding motif (Figure 1), which according to our analysis is rather well conserved among eutherian mammals (Supplementary file S1). However, whereas the PMEPA1-001 transcript open reading frame and a part of the intergenic promoter region are highly conserved through evolution, this seems to be true to a lesser degree for NKILA equivalent transcripts (Supplementary file S1 and discussion therein). So, from the standpoint of evolution, a logical hypothesis for the function of the seemingly younger NKILA is its possible interference with PMEPA1 expression4,5.
PMEPA1 expression is strongly enhanced by TGF-β6,7, something which agrees with the two SMAD binding element motifs that we found conserved in its promoter (Supplementary file S1). PMEPA1 function is not well understood, but it is believed to encode a transmembrane protein that with its cytoplasmic domain can bind SMAD proteins and can positively affect activation of Akt7. Signaling pathways involving Akt and NF-κB are known to converge8, which might be relevant for a possible indirect effect of NKILA through PMEPA1 on NF-κB functions. PMEPA1 levels were reported to be high in invasive MDA-MB-231 breast cancer cells, and low in non-invasive MCF-7 and T47D breast cancer cells, agreeing with observations for aggressive versus non-aggressive tumors9. This is exactly opposite to the expression pattern observed by Liu et al. for NKILA in these cell lines and among tumors. PMEPA1 knockdown has been found to be able to attenuate growth and motility of MDA-MB-231 breast cancer cells9, which is interesting since Liu et al.1 found that forced NKILA expression (which might knockdown PMEPA1 expression) in MDA-MK-231 cells achieves similar effects. Consistent with these findings is the observation that high PMEPA1 expression in breast cancer is associated with poor patient prognosis7, and high NKILA expression with better patient prognosis1. Although there are also published PMEPA1 reports which are harder to reconcile with such a model (e.g. reference 10), and which are hard for us to validate, at least the above selected set of data suggests that NKILA has a negative effect on PMEPA1 function. More research on both NKILA and PMEPA1 will be necessary before conclusions can be made, but for now the possibility that NKILA can downregulate PMEPA1 expression appears to be a reasonable model4,5.
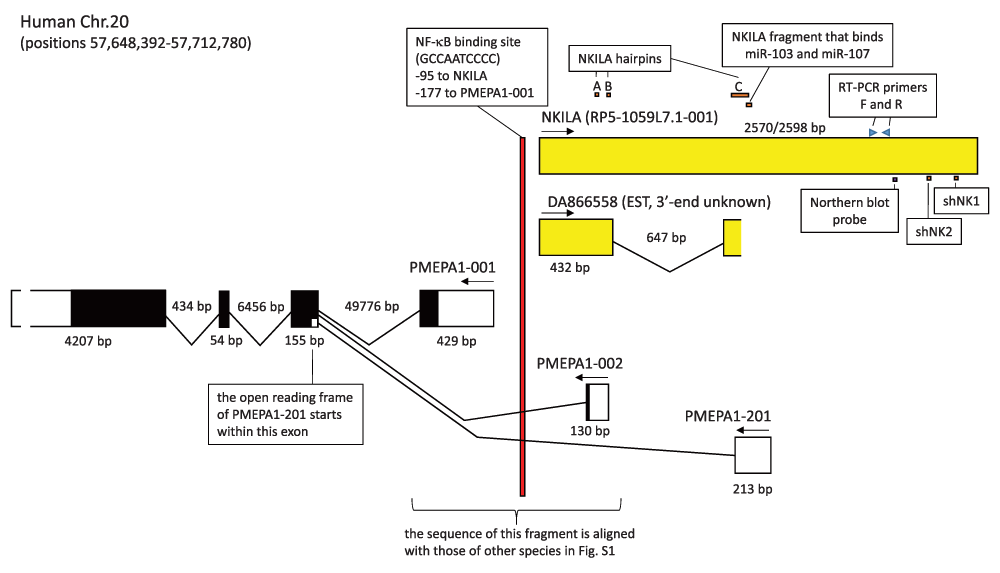
The figure summarizes several data from the study by Liu et al. for NKILA and its promoter, while also showing overlapping transcripts that were neglected in that study. The NKILA transcript identified by Liu et al. roughly corresponds with transcript RP5-1059L7.1-001 as summarized in the GRCh38.p2 dataset of the Ensembl database (http://www.ensembl.org/index.html). GenBank accession DA866558 (RP5-1059L7.1-002 in Ensembl) contains an expressed sequence tag (EST) which represents the 5’ end of a spliced transcript and for which the 3’ end is not known. The depicted summary of the PMEAP1 transcripts -001, -002 and -201, is derived from the Ensembl database and agrees with GenBank reports; for additional variations of PMEAP1 transcripts we refer to the Ensembl database. Exons are indicated by boxes, with protein coding regions in black. The 3’ UTR of PMEPA1 is not drawn in correct proportion to the other exon regions. Arrows indicate the direction of transcription, and genomic regions are measured in basepairs. The figure also shows from the Liu et al. report the positions of the NF-κB binding promoter element, the NKILA hairpin-prone regions, the NKILA region that binds miR-103 and -107, the NKILA binding sites for the Northern blot probe and RT-PCR primers, and the shNK1 and shNK2 regions from which sequences were derived for cloning into shRNA constructs.
Liu et al.1 concluded that NKILA interferes with pathways that involve NF-κB function, and we feel that in essence this conclusion can be believably deduced from their abundant experimental data. However, mechanistically Liu et al. only consider a direct interaction with NF-κB components and fail to consider an indirect effect through PMEPA1 regulation. Liu et al.1 did find direct binding between the NF-κB component p65 and NKILA, but whether this can be considered as evidence of physiological specificity of NKILA for NF-κB is questionable. When they analyzed proteins that they could pull down with NKILA they only compared different NF-κB pathway components using Western blot analysis1. In addition, when they quantified NKILA by RT-PCR on genetic material that co-precipitated with NF-κB factors, they did not exclude the possibility that they might be measuring genomic NKILA DNA1.
Liu et al.1 mapped the NKILA interaction with p65 to hairpin-prone regions A and B, and showed that the hairpin-prone region C can interact with NF-κB pathway factor IκBα (for hairpin-prone region locations see Figure 1). By mutation analysis, Liu et al.1 found that all these three hairpin-prone regions are important for the inhibitory effect of NKILA on NF-κB activity. Although the relevance of this overlap is not clear, we point out that all three identified hairpin-prone regions, and also the region which Liu et al.1 found to confer sensitivity to miRNA induced degradation, overlap with known PMEPA1 transcript regions (Figure 1).
As an additional remark, we would like to state that the somewhat discussable manner in the way Liu et al.1 performed or described some of their experiments (see our comments in Supplementary file S2) does not help to convey the image of a study which is solid in its quantitative aspects. However, despite our criticism, it is only fair to state here that according to our judgement the very elaborate study by Liu et al.1 believably shows (1) how NKILA can bind (in vitro) to NF-κB, (2) that NKILA can interfere with functions that involve NF-κB pathways, and (3) that low NKILA expression predicts poor clinical outcome in patients with breast cancer. But they should mend the open ends, which means providing more evidence of the specificity of the NKILA binding to NF-κB, and to take the possible effects of NKILA through regulation of PMEPA1 into consideration. In regard to the more general claim by Liu et al.1 that there exists “a class of lncRNAs that regulate signal transduction at post-translational level”, we believe as before11 that such a conclusion needs more evidence than currently has been presented. We hope that our present discussion leads to an increased interest in the PMEPA1-NKILA locus, because whatever mechanism may be correct, Liu et al. did provide evidence that this locus is clinically important in breast cancer.
JMD initiated the study. DBA and JMD performed the analysis. JMD wrote the article with help of DBA. Both authors critically edited the correspondence and agreed to the final content.
Supplementary file S1.
Alignment of PMEPA1-NKILA promoter region sequences of representative animals and deduced PMEPA1 amino acid sequences for the species compared.
Click here to access the data. http://dx.doi.org/10.5256/f1000research.6400.s45984
Supplementary file S2.
List of issues regarding the Liu et al. (2015). Cancer Cell 27, 370–381 paper which in our opinion need attention.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 22 Apr 15 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Thank you for your reply. The best way to address the possibility that NKILA may regulate PMEPA1 expression is by experiments indeed. Because of the timing of your ... Continue reading Dear Dr. Liu,
Thank you for your reply. The best way to address the possibility that NKILA may regulate PMEPA1 expression is by experiments indeed. Because of the timing of your data presentation I am a bit suspicious of them, but I'll give you the benefit of the doubt. Probably there are some remaining questions as how the shared PMEPA1-NKILA promoter region works, e.g. in TGF-β vs TNFα stimulation, and in MDA-MB-231 vs MCF7 cells. It may be a good choice to look for a cooperation with Dr. Saikumar. If both of your groups would find the same thing on expression regulation, it would be truly believable, and both of your groups would benefit from the knowledge. Several journals like for example F1000Research accept articles that are descriptive in nature, so you wouldn't even have to determine the precise regulatory mechanisms. The above is just a suggestion, but, especially in regard to breast cancer, NKILA and PMEPA1 research seem to belong together, and it seems to be a solid way to regain the thrust of your readers.
Just a small lay-out comment regarding the figure that you added to your reply is that you probably should use white as the color for the Fig.1b rods, since now it looks as if they represent samples after TNFα stimulation (and they don't, right?).
If you would like to better nail the in vivo specificity of NKILA to NF-kB binding, you might be interested in a recent paper in which they used mass spectrometry analysis for proteins bound by lncRNA (McHugh et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015 Apr 27. [Epub ahead of print] ).
As before, I believe that your study shouldn't have been published the way you did, but also believe that if you would add the right experiments your important message may survive.
Sincerely,
also on behalf of David B. Alexander,
Johannes M. Dijkstra
Thank you for your reply. The best way to address the possibility that NKILA may regulate PMEPA1 expression is by experiments indeed. Because of the timing of your data presentation I am a bit suspicious of them, but I'll give you the benefit of the doubt. Probably there are some remaining questions as how the shared PMEPA1-NKILA promoter region works, e.g. in TGF-β vs TNFα stimulation, and in MDA-MB-231 vs MCF7 cells. It may be a good choice to look for a cooperation with Dr. Saikumar. If both of your groups would find the same thing on expression regulation, it would be truly believable, and both of your groups would benefit from the knowledge. Several journals like for example F1000Research accept articles that are descriptive in nature, so you wouldn't even have to determine the precise regulatory mechanisms. The above is just a suggestion, but, especially in regard to breast cancer, NKILA and PMEPA1 research seem to belong together, and it seems to be a solid way to regain the thrust of your readers.
Just a small lay-out comment regarding the figure that you added to your reply is that you probably should use white as the color for the Fig.1b rods, since now it looks as if they represent samples after TNFα stimulation (and they don't, right?).
If you would like to better nail the in vivo specificity of NKILA to NF-kB binding, you might be interested in a recent paper in which they used mass spectrometry analysis for proteins bound by lncRNA (McHugh et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015 Apr 27. [Epub ahead of print] ).
As before, I believe that your study shouldn't have been published the way you did, but also believe that if you would add the right experiments your important message may survive.
Sincerely,
also on behalf of David B. Alexander,
Johannes M. Dijkstra