Keywords
Salmonella typhimurium, cell growth, outer membrane, membrane protein, YejM, YejL, cardiolipin
Salmonella typhimurium, cell growth, outer membrane, membrane protein, YejM, YejL, cardiolipin
Salmonella typhimurium is a Gram-negative bacterium responsible for over 35% of all foodborne illness-related hospitalizations in the United States (Painter et al., 2013). S. typhimurium possesses an additional outer membrane (OM) with an asymmetric lipid composition, that serves as a barrier to the environment allowing its survival and replication within host tissues (Dalebroux & Miller, 2014; Needham & Trent, 2013; Pagès et al., 2008). Mechanisms for the transport and assembly of OM lipopolysaccharides, proteins, and exopolysaccharides have been defined (Dong et al., 2006; Dong et al., 2014; Hagan et al., 2011; Whitfield & Trent, 2014); however, the transport of glycerophospholipids across the periplasm and insertion into the inner-leaflet of the OM is not well understood.
Interestingly, increased levels of cardiolipin in S. typhimurium OM were observed upon PhoPQ regulator activation (Dalebroux et al., 2014). Recently the inner membrane protein YejM was shown to bind cardiolipin and be involved in OM formation (Dalebroux et al., 2015). Furthermore, YejM is known to be an essential gene in E. coli and was shown to be involved in intrinsic multidrug resistance (De Lay & Cronan, 2008; Duo et al., 2008). YejM is comprised of 586 amino acids forming five predicted N-terminal transmembrane helices, followed by an arginine-rich periplasmic random coil linker region, and a C-terminal periplasmic domain (Figure 1A), and it was shown that YejM associates as a tetramer in solution (Dalebroux et al., 2015). The arginine-rich linker region and periplasmic globular domain of YejM were shown to bind cardiolipin and are required for OM remodeling and cell growth (Dalebroux et al., 2015). The molecular mechanism of the interplay between PhoPQ system and YejM, and how cardiolipin molecules are transported to the OM, need further structural and functional investigation.
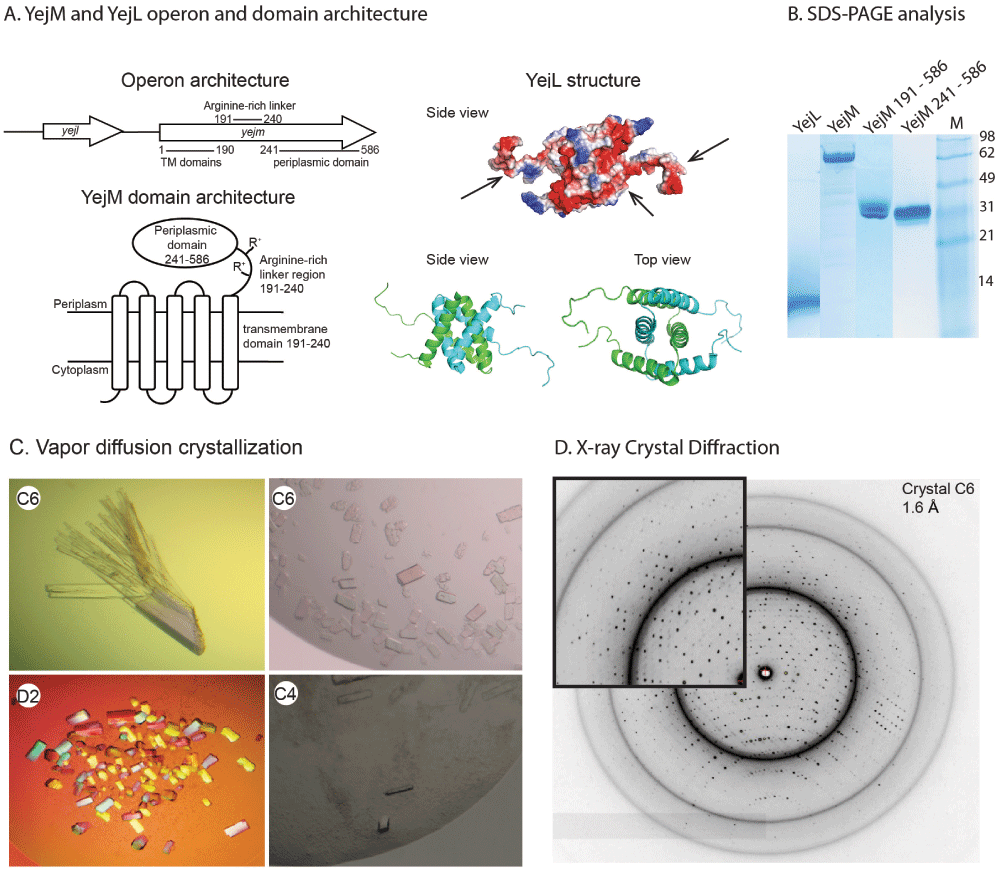
A) Operon architecture containing genes yejl and yejm, YejM protein domain architecture, YejL structure (PDB ID: 2JRX) showing electrostatic surface pattern, black arrows indicating negative charged areas, and in cartoon representation the two intertwined dimer (chain A in green and chain B in blue) forming a four-helix bundle B) SDS-PAGE analysis of purified protein samples. C) Crystals of YejM241 under various conditions. D) Diffraction image of YejM241 crystal from condition C6.
Interestingly the gene yejl is localized upstream of yejm on the same operon (Figure 1A), and encodes for the 8.5 kDa protein, YejL, that forms a homodimer (pdb ID: 2JRX) (Figure 1A). The function of YejL is not known; however based on the co-location on the operon and YejL’s negatively charged areas shown in the homodimer NMR structure, we hypothesize that YejL is a ligand to YejM. We further hypothesize that YejL binds to the same site on YejM as cardiolipin and therefore may be a regulator of YejM-mediated cardiolipin transport.
Here we report how YejM was engineered to facilitate crystal growth and present successful crystallization conditions with preliminary X-ray diffraction analysis. We further report for the first time that YejL is a ligand of YejM. Future successful structure determination of YejM alone and in complex with YejL will help us to understand cardiolipin transport to the OM and may lead to new drug targets inhibiting the pathogenic properties of S. typhimurium.
We purified full-length YejM and periplasmic constructs YejM191-586 as described in (Dalebroux et al., 2015). The original construct YejM191-586 failed to crystallize and showed a degradation product after electrophoresis in the SDS polyacrylamide gel (Figure 1B, middle lane). To prevent degradation, reduce flexible protein parts and remove positively charged arginine clusters, we deleted the linker region A191 to E240 in the YejM191-586 construct, resulting in YejM241-586. These modifications were made to increase the chance of crystal growth. Initial crystals of YejM241-586 appeared after one week incubation at 18°C under different conditions; e.g. needle clusters in 2.8 M sodium acetate trihydrate pH 7.0, 0.1 M BIS-TRIS propane pH 7.0 (Hampton SaltRx condition A2, Hampton Research), needle clusters in 2.8 M sodium acetate (Hampton Index condition B12, Hampton Research), and rhombohedral crystals appeared in 3.5 M sodium formate pH 7.0 (Hampton Index HR conditions C1, Hampton Research). These early hit crystals diffracted poorly, only up to 6Å (data not shown) and optimization and up-scaling of the crystallization setup from a 96 well format to a 24 well format did not improve the diffraction quality. Further screening using (Hampton PEGRx HT screen, Hampton Research) resulted in new crystal forms grown in condition C6 (0.1 M HEPES pH 7.5, 12% w/v polyethylene glycol 3,350) and C4 (0.1 M Citric acid pH 3.5, 25% w/v polyethylene glycol 3,350). Screening around these two conditions led to crystals in a condition consisting of 0.1 M citric acid pH 4, 18% w/v polyethylene glycol 3,350 (Figure 1C). YejM241-586 crystals grown in condition C6 diffracted well, up to 1.6 Å (Figure 1D). Data indexing and scaling with XDS (Kabsch, 2010) and further analysis with AIMLESS (Evans, 2006) resulted in a data set up to 1.8Å resolution and good overall statistics (Table 1).
The gene yejl is localized upstream on the same operon as yejm. yejl encodes for a small (8.5 kDa) protein, YejL, that forms an intertwined four-helix bundle (PDB code: 2JRX) and has negatively charged regions located at the “tail” ends of the four-helix bundle (Figure 1A). The biological function of YejL is unknown, however, its presence on the same operon as YejM suggests that it may interact with YejM and function as a regulatory element. More specifically, we hypothesize that the negatively charged regions of YejL could be involved in specific interaction with the positively charged arginine-rich linker region of YejM. Notably, this arginine-rich linker region is proposed to bind cardiolipin and may be a crucial region for cardiolipin translocation by YejM (Dalebroux et al., 2015). To test our hypothesis that YejL is a ligand of YejM, we purified and mixed both proteins in different stoichiometric ratio and conducted Blue Native PAGE (BNE) and size-exclusion chromatography (SEC) for detection of population of higher molecular weight oligomers/complexes in the mixtures. Our SEC experiments showed that, increasing concentrations of YejL fraction into YejM191-586 or YejM241-586 fractions, respectively (Figure 2A) resulted in a shift in the apparent molecular weight that suggested formation of a higher molecular weight complex. In BNE, we observed a clear shift in position of YejL towards YejM191-586 and YejM241-586, which indicated binding between YejL and YejM191-586 or YejM241-586, respectively (Figure 2B). Our data do not show an apparent difference between YejL binding to YejM191-586 that includes the arginine-rich linker region, and YejM241-586 that lacks the linker region. Further analysis using isothermal titration calorimetry and/or microscale thermophoresis will be used to determine the exact binding affinity and potential changes thereof between the two YejM constructs and YejL.
Here we report successful crystallization using protein engineered specifically to enable crystal growth of YejM. Our initial X-ray data analysis of YejM241-586 crystals suggests a dimer assembly of the periplasmic domain. Therefore the membrane domain is very likely needed to form the YejM tetramer. We will use the current 1.8Å dataset (Table 1) for structure determination. We also aim to solve the co-crystal structure of YejL and/or cardiolipin bound to YejM. Ultimately these structures will help in understanding: i) where and how YejL and/or cardiolipin bind to YejM, ii) the role of YejL, iii) whether YejM’s architecture is that of a transporter or channel, and iv) the molecular mechanism of cardiolipin translocation to the OM of S. typhimurium.
Initial clones of full-length YejM 1-586 (YejM) (Uniprot ID P40709) in pBAD24 and the periplasmic domain of YejM 191-586 in pET28a plasmid are described in (Dalebroux et al., 2015). We used forward primer YejM241-586 5'-ccgcgcggcagccatatggctagcgcggtctccgttcagtacccg- 3' and reverse primer YejM241-586 5'-gcgggtactgaacggagaccgcgctagccatatggctgccgcgcgg- 3' to create a shorter construct of the periplasmic domain lacking the linker region resulting in YejM 241-586. Purification of YejM, YejM 191-586, and YejM 241-586 was performed as described previously (Dalebroux et al., 2015). Samples used for subsequent crystallization experiments were further purified by SEC using a Superose 6 increase 10/300 GL column (GE Healthcare) in buffer containing 50 mM Tris pH 8.0, and 150 mM NaCl. SEC buffer for YejM contained 20 mM HEPES, pH 7.5, 150 mM NaCl, and 0.02% Dodecyl-β-D-Maltopyranoside (DDM). The concentration of DDM was kept right above the critical micelle concentration throughout all subsequent experiments. YejM and YejM241-586 SEC peak fractions were pooled and concentrated with 30 kDa NMWL centricon (Millipore) to 12 mg/ml and up to 50 mg/ml, respectively. The purity of the samples was judged by polyacrylamide gel electrophoresis (Figure 1B).
YejL construct (ID ER309-21.7), an E. coli homolog, was obtained from the Northeast Structural Genomics (NESG) consortium (http://www.nesg.org) with the vector ID: pET21_NESG. The pET21-YejL plasmid was transformed into E. coli BL21(DE3) (Novagen) competent cells. Overnight saturated cultures of transformed bacteria in Luria Bertani (LB) medium (EMD Chemicals Inc.) with 50µg/ml ampicillin (Dot Scientific Inc.) were grown at 37°C and diluted 1:200 in fresh Terrific Broth (TB) medium (Dot Scientific Inc.) with 50µg/ml ampicillin and further grown at 37°C at 200 rpm. The cultures were induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) (Teknova) when OD600 reached 0.5–0.8. The expression was carried out at 37°C for three hours. All further steps were performed at 4°C unless noted otherwise. Cells were harvested by centrifugation and resuspended in a buffer containing 50 mM Tris, pH 8.0, 150 mM NaCl, and 5 mM MgCl2. 10µg/ml DNAse (Sigma Aldrich), 1 mg/ml lyzosyme (Sigma Aldrich), protease inhibitor cocktail (Roche), and 0.1 mM Phenylmethane sulfonyl fluoride (PMSF) (Dot Scientific Inc.) were freshly added to the resuspended cells. Cells were lysed by passing several times through a microfluidizer (Divtech Equipment Company) and the cell debris was removed by centrifugation.
The clear cell lysate was bound to Ni-NTA resin (Qiagen) in batch mode and incubated on a rotarod at 4°C for 30 – 60 minutes. The slurry was loaded into a column and the flow through was collected under gravity. The column was washed first with 30 column volumes of Wash buffer 1 (25mM Tris pH, 8.0, 300mM NaCl and 25mM Imidazole); then with two column volumes of Wash buffer 2 (25mM Tris pH 7.5–8, 150mM NaCl, 75mM Imidazole). The bound protein was eluted from the column with elution buffer (25 mM Tris, pH 8.0, 150 mM NaCl, and 300 mM Imidazole) in 200–500µl increments. Elutions were monitored for protein content by Bradford test. YejL was further purified by SEC using a Superose 6 increase 10/300 GL column (GE Healthcare) equilibrated in a buffer containing 50 mM Tris, pH 8.0 and 150 mM NaCl. Peak fractions of YejL were consolidated and concentrated using 3 kDa NMWL centricon (Millipore).
Vapor diffusion crystallization of YejM 241-586 (5–50 mg/ml) was set up using 96-well crystallization plates (Hampton Research) with a Phoenix robot (ARI). Various sparse matrix screens were used to set up sitting drops with a drop sizes between 500 nl to 1 µl. Crystallization plates were incubated at 20°C and monitored for crystal growth in a MinstelTM HT crystal imaging and detection tower (Rigaku). Optimal crystal growth was obtained at a protein concentration of 4mg/ml.
10 µM of YejM191-586 or YejM241-586 was mixed with varying concentrations (2.5, 5, 10, and 15 µM) of YejL separately. Samples were incubated at 4°C for minimum 1 hr, mixed with sample buffer (5% Coomassie Brillant Blue G250, 100mM Bis-Tris pH 7.5, 0.5M 6-aminocaproacid (Biorad) for a total individual sample volume of 30μL. The samples were loaded on Mini-protean TGX Any-kD precast gels (Biorad) and electrophoresis was carried out at 4°C at 100 V for 60–90 minutes. The gels were destained in a solution containing 5% ethanol and 7.5% acetic acid.
10 µM of YejM191-586 and YejM241-586 were each mixed with varying concentrations (2.5, 5, 10, and 15 µM) of YejL separately. Each protein mixture was incubated overnight at 4°C and subsequently purified by size-exclusion chromatography using Superose 6 Increase 10/300 GL gel-filtration column (GE Healthcare) equilibrated with 50 mM Tris pH 8.0, 150 mM NaCl. The UV absorption profiles at 280 nm of each run were normalized and compared.
Crystals were harvested using Litho loops (Molecular Dimensions) and Nylon loops (Hampton Research), submerged into paraffin and blotted until no phase separation was visible between paraffin and the excess crystallization solvent. Diffraction data of YejM241-586 crystals were collected at the Advanced Light Source beamline 4.2.2 in Berkeley CA at 100K, using an oscillation of 0.1–0.2° per image. Diffraction data were processed using iMosflm (Powell et al., 2013) or XDS (Kabsch, 2010) and scaled with Scala (Evans, 2006).
Raw diffraction data images were uploaded to the Coherent X-ray Imaging Data Bank (http://cxidb.org/id-42.html) and are available under CXIDB ID 42, DOI 10.11577/1252489 (Gabale et al., 2016).
U.G. expressed and purified proteins, performed SEC and BNE, analyzed data and wrote manuscript, G.Q and E.R expressed, purified, crystallized and performed BNE, S.R. expressed, purified, crystallized, collected and analyzed data, designed experiments and wrote manuscript.
We thank: Prof. Samuel I. Miller and Dr. Zachary D. Dalebroux for constructs of full-length YejM and periplasmic domain of YejM191-586, Richard Pfuetzner for stimulating discussions, Jonathan T. Siler and Jennifer Wong for help with initial protein purification and crystallization experiments, Dr. Ardian Soca Wibowo for his excellent service at the macromolecular crystallization facility at Indiana University Bloomington, and Dr. Jay Nix for his excellent support at Molecular Biology Consortium Beamline 4.2.2 at the Advanced Light Source (Berkeley, CA).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
References
1. Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ressl S, et al.: Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence.Cell Host Microbe. 2015; 17 (4): 441-51 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 11 Dec 17 |
read | |
|
Version 1 02 Jun 16 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)