Keywords
CHEK2, CHK2, cancer, Latin America, databases, mutations, CHEK2*1100delC, genomics
CHEK2, CHK2, cancer, Latin America, databases, mutations, CHEK2*1100delC, genomics
CHEK2 (Checkpoint Kinase 2) (OMIM +604373) encodes CHK2 a serine/threonine kinase that is the human homolog of Saccharomyces cerevisiae RAD53 and Schizosaccharomyces pombe CDS11. In mammalian cells, ATM activates CHK2 in response to ionizing radiation through phosphorylation. This leads to a variety of cellular responses, such as cell cycle checkpoint activation2, where CHK2 is involved in maintaining the G1/S and G2/M checkpoints by phosphorylation of CDC25A, CDC25C and p533 and in the repair of double-strand DNA breaks via homologous recombination (HR) through phosphorylation of BRCA14 and BRCA25. CHK2 is also involved in the induction of p53-dependent apoptosis through phosphorylation of p53 on Ser206, and, in a p53-independent manner, via phosphorylation of PML and E2F13. These responses prevent damaged cells from going through the cell cycle or proliferating. CHK2 also plays an important role during mitosis by maintaining chromosomal stability7.
CHEK2 c.1000delC, a truncating mutation in exon 10 that abolishes kinase activity of the protein, was the first mutation being reported for this gene and was found in a woman with breast cancer and family history of Li-Fraumeni syndrome-28. The role of this mutation in breast cancer was confirmed by Meijers-Heijboer et al.9 and in several other studies10–22. Based on these studies, CHEK2 has been proposed as a moderate penetrance breast cancer susceptibility gene9 and mutations in this gene are associated with almost a 3-fold increase in the risk of breast cancer in women and a 10-fold increase in the risk of breast cancer in men23.
Given the role of CHEK2 in maintaining genomic stability and the fact that the CHEK2 protein is expressed in a wide range of tissues, it was not surprising that alterations in this protein were found in other cancers, including glioblastoma, ovarian, prostate, colorectal, gastric, thyroid, and lung cancer18,24–28. The studies in CHEK2 included individuals mainly from the United States and Europe while Latin American individuals were underrepresented. In order to infer the role of the CHEK2 gene in the cancer etiology in the Latin American population we compiled mutations in the CHEK2 gene registered in genomics data repositories and the literature, that had been reported in this population.
Mutations in CHEK2 were identified in The Exome Aggregation Consortium (ExaC, RRID:SCR_004068, http://exac.broadinstitute.org/)29 browser, the Cancer Genome Atlas (TCGA, RRID:SCR_003193)30 data sets extracted from the cBioPortal for Cancer Genomics (RRID:SCR_014555, http://www.cbioportal.org/)31, and The International Cancer Genome Consortium (ICGC) (http://icgc.org/)32. From the GWAS catalog (RRID:SCR_012745, https://www.ebi.ac.uk/gwas/)33 a list of SNPs mapped to CHEK2 and associated with a disease was also downloaded. Data obtained from cell line studies was not included.
ICGC, the cBioportal and ExAc use prediction tools to assess functional impact of non-synonymous (SO term: missense_variant) somatic mutations on protein coding genes. ICGC uses FatHMM (http://fathmm.biocompute.org.uk/)34, Mutation Assessor (RRID:SCR_005762)35 and SIFT (RID:SCR_012813)36 to compute functional impact scores and assign impact categories (High, Medium, Low and Unknown). The cBioPortal uses Mutation Assessor and reports the same impact categories. We used those functional impact categories to filter the mutations and extract possible pathogenic mutations by selecting only high and medium impact mutations and nonsense alterations. The percentage of mutations in CHEK2 per cancer study and the percentage of cases altered per cancer type was also calculated. The filter used for the ExAC information was based on the annotation of possible damaging and deleterious mutations made by two in silico tools: Polyphen2 (RID:SCR_013200)37 and SIFT36. The assessment of stop gained, splice site disrupting and frameshift variants was made through Loss of Function Transcript Effect Estimator (LOFTEE), a plugin of the Ensembl Variant Effect Predictor (VEP) (RRID SCR_007931)38. The Latino annotation was examined in the databases that reported ethnicity data; this search was done before filtering the datasets, with the purpose to report all genetic alterations found in Latin American populations.
The plots were generated with R version 3.3.1 (RRID:SCR_001905)39.
In order to include all the studies identifying CHEK2 gene mutations in Latin America, a deep search of literature was conducted by using the terms “CHEK2”, “CHEK2 Latin America”, and “CHEK2 cancer” in electronic academic literature search engines. PUBMED (RRID:SCR_004846) was the relevant database used followed by Google Scholar (RRID:SCR_008878). References of the retrieved articles were also screened for relevant studies. This search strategy was performed iteratively up to and including 10 October 2016.
The complete list of mutations in CHEK2 reported in the cBioPortal and ICGC, before applying filters, are available in Dataset 1 and Dataset 2, respectively.
cBioPortal. The available data sets consisted of 147 studies that included only cancer samples. Mutations in CHEK2 were reported in 39 out of the 147 studies. Before applying filters, cholangiocarcinoma (8.6%), uterine carcinosarcoma (7.0%), and colorectal adenocarcinoma (6.9%) were the types of cancer that showed the higher number of cases (Figure 1); meanwhile, breast, colorectal and non-small cell lung cancer (NSCLC) had more mutations in CHEK2 than other cancer types (Figure 2).
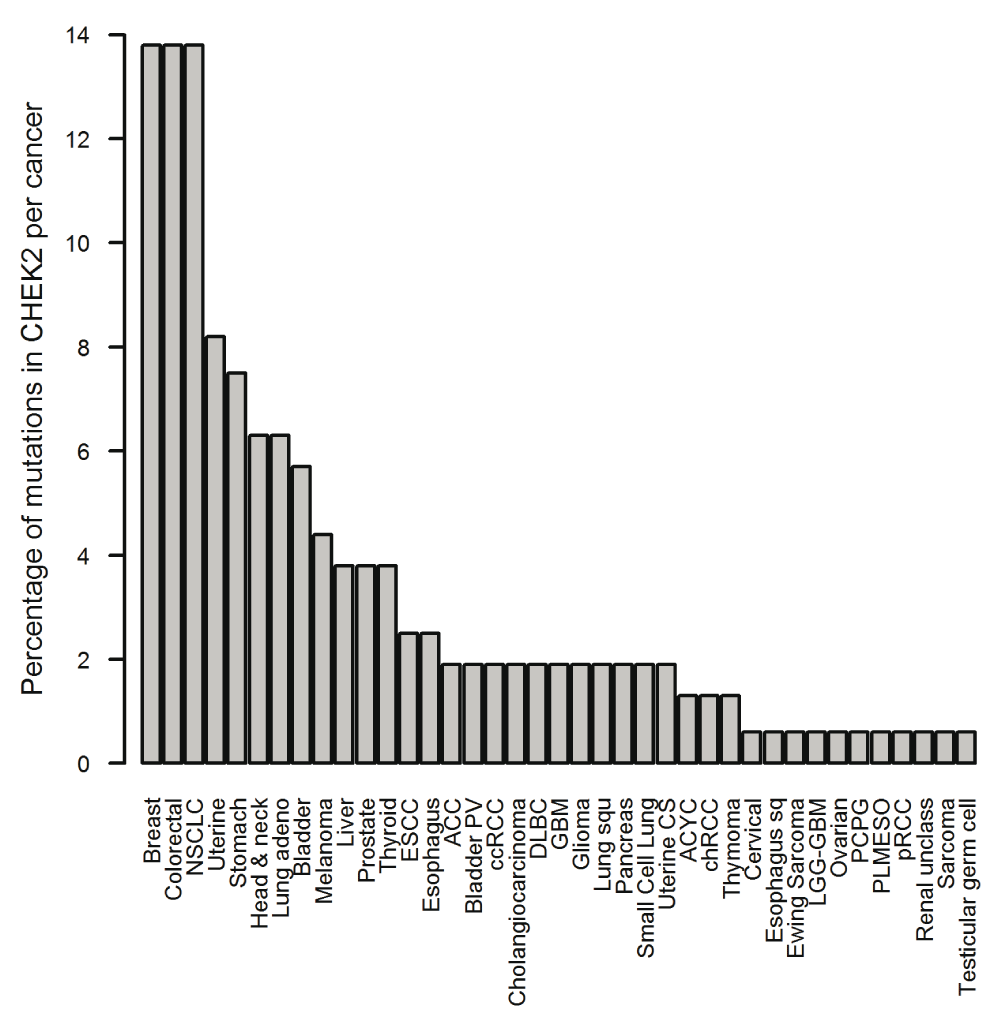
The X axis shows the type of cancer in which at least one case has a mutation in CHECK2, the Y axis indicates the percentage of cases per study that have mutations in CHECK2 (source: cBioPortal).
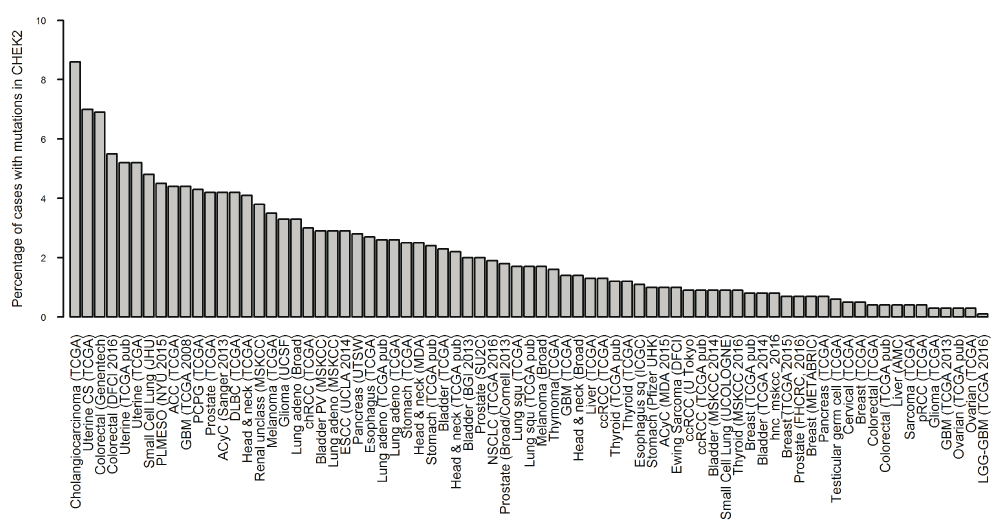
The X axis shows the type of cancer in which at least one mutation in CHECK2 was identified, the Y axis indicates the percentage of mutations in CHEK2 per cancer type (source: cBioPortal). n unique mutations = 159. Synonymous mutations are not included in the cBioPortal database.
Using the Mutation Assessor from cBioPortal, we filtered out mutations labeled to have neutral and low impact. In Table 1 we are reporting the mutations with high and medium impact and also nonsense mutations and frameshifts. Table 1 shows the 78 mutations that remained after the filtering process, 38 of which were classified as with high impact. 51.2% of mutations were missense mutations, 20.5% were frameshift mutations, 19.2% were nonsense mutations and 9% were in splice sites. The type of cancer with most mutations (13/78) was breast cancer, followed by uterine, lung, and colorectal cancer. The rest of cancer types had six or less mutations. The most frequent mutation was E321* reported in three patients with uterine cancer.
Before filtering the mutations found in the cBioPortal we identified Latino individuals with the ethnicity data obtained from the TCGA clinical data available at the NCI's Genomic Data Commons portal (GDC, RRID:SCR_014514, https://gdc-portal.nci.nih.gov/) (Table 2). Two patients with three mutations in the gene were found. One of the samples was a Latino patient from the head and neck squamous cell carcinoma cohort (HNSC); this patient carries the neutral variant K373E. Because this is a neutral variant it was not included in Table 2. The second Latino patient was part of the diffuse large B-cell lymphoma (DLBC) cohort; this patient carries a frameshift and a nonsense mutation.
A total of 279 mutations including up- and down-stream mutations were reported in 185 donors. From this number, seven mutations are predicted to have high impact (Table 3). For the Latin American population in ICGC, the Brazilian melanoma study (SKCA-BR) reported four mutations inside the gene, one of them with high impact (Table 2 and Table 3).
*Depending of transcript. All mutations are single base substitutions. MELA-AU: melanoma, Australia. BRCA-EU: breast ER+ and HER- cancer, European Union. ESAD-UK: esophageal adenocarcinoma, United Kingdom. SKCA-BR: skin adenocarcinoma, Brazil. LINC-JP: liver cancer, Japan. BRCA-FR: breast cancer, France.
A total of 742 mutations for the CHEK2 gene were reported in this database and 132 of them were present in the Latino population before filters (Dataset 3). After applying the filter of possibly damaging and deleterious alterations, 23 mutations in the Latino population were left. In this group the mutation p.Leu279Pro was the most frequent (0.003112). CHEK2 c.1100delC (p.Thr410MetfsTer15*), the most interrogated mutation in CHEK2, was found in two samples (Table 2).
Mutations rs132390-C and rs17879961-A mapped to or near CHEK2 were associated in European populations with breast and lung cancer, respectively. Mutations rs4822983-T and rs2239815-T were associated with esophageal squamous cell carcinoma in individuals with Han Chinese ancestry. In addition, in a Han Chinese cohort of esophageal and gastric cancer the mutation rs738722-T was also associated with those cancers (Dataset 4).
In total, we found nine studies in which mutations in CHEK2 were evaluated in Latino populations. Two of these studies were international and included Latin American cancer patients10,22 and the other six studies were country-based. The country in which most studies have been performed was Brazil with four studies40–43. In Argentina44, Chile45, and Mexico46 one study per country was identified. In eight out of the nine studies, the presence of variants in CHEK2 was interrogated in breast cancer patients. Only one study used samples of patients with hereditary breast and colorectal cancer. The mutation most frequently evaluated in these investigations was c.1100delC (in six studies); while other two studies42,44 interrogated the other two most frequent mutations in the CHEK2 gene (c.470T>C and c.444+IG>A) in addition to c.1100delC. Additionally, Chaudury et al. performed a complete sequencing of the gene and found a different mutation, c.478A>G (p.Arg160Gly)46. Table 4 shows the Latin American studies that reported the presence of mutations in CHEK2 mutations and their frequency.
A search in cancer genomics data repositories and the literature was performed to identify mutations in CHEK2 in different cancer types, with specific emphasis on mutations found in Latino American populations. The database with the most number of mutations reported in CHEK2 for Latino populations was ExAC with 132 mutations, followed by ICGC with four mutations, and TCGA with three mutations. After filtering 30 mutations with high and medium impact according to the databases functional impact categories were kept: seventeen missense, eight ‘stop gain’ mutations, one frameshift mutation, two mutations in the 5’UTR, and two mutations in splice donor sites of CHEK2. These mutations included the most analyzed mutation of CHEK2, c.1100delC (p.Thr367Metfs) (Table 2).
Worldwide, according to our findings in the ICGC and TCGA databases, CHEK2 mutations were reported in 23 cancer types, while in the Latin American population CHEK2 mutations were only found in head and neck cancer, lymphoma and melanoma. In this context, it is important to highlight, that Latino populations have been underrepresented in other worldwide studies. As shown in Dataset 4, the cohorts of TCGA are biased toward the inclusion of white individuals and individuals from other ethnicities are underrepresented. The same was observed in ICGC in which only a Latin American cohort from Brazil was available for our analysis. Regarding the data found in our literature review, CHEK2 has only been studied in the Latin American population in breast and colorectal cancer.
In the ExAC repository, the mutations c.1100delC and c.478A>G were found two times and one time, respectively, in the Latino population (Dataset 3). In TCGA, c.1100delC was found in a patient with breast cancer but information about its ethnicity was not available (Table 1). Up to now, only nine studies evaluating mutations in CHEK2 have been performed in Latin America and only six of them found mutations in the gene, five studies found the c.1100delC mutation and one found the c.478A>G (p.Arg160Gly)10,22,40,43,46. Two mutations, c.1100delC and c.478A>G, were classified in the ClinVar archive (https://www.ncbi.nlm.nih.gov/clinvar/) as pathogenic and likely pathogenic, respectively. These mutations are the only ones in common with the mutations found in genomics data repositories.
Although c.1100delC is the CHEK2 mutation most evaluated in the Latin American population, it should be noted that its frequency, seen from literature reports and data repositories, is rather low. Because the highest frequency of this mutation is found in populations from the Northern and Western Europe, c.1100delC is proposed as an allele with population gradient, which originated in these populations and its frequency decreases as you get to the southern regions of Europe (Basque Country, Spain, and Italy)47. Taking into account the European genetic component of Latin American populations, it is expected that if the frequency of c.1100delC is low in the Spanish population, in our mixed populations the frequency would be even lower.
Because cancer types other than breast and colorectal cancer, such as uterine, lung, bladder and head and neck cancer, presented mutations in CHEK2 in several populations, it is relevant to focus the search for mutations in these types of cancer in the Latin American populations. Additionally, the interrogation of CHEK2 mutations in the Latin American population has been focused mainly on the c.1100delC mutation, but the data obtained from the ExAC database showed that in Latin American samples there are 23 germline mutations (Table 2) that could generate cancer susceptibility. It would therefore be important to examine the frequencies of these mutations in the Latin American population and its association with the development of cancer.
This study has limitations; for example, information about race and ethnicity was not available for at least 28 studies in the cBioPortal, and consequently some Latinos may be hidden in those studies. Thus, the small number of Latinos included in the genomics data repositories could be a reason why we have found a small number of mutations in CHEK2 in this population. It is important to highlight that the use of different transcripts for reporting mutations makes the correlation between mutations found in different studies laborious.
This study presents a compilation of mutations in CHEK2 with high impact in different cancer types in White, Hispanic and other populations. We also showed the necessity of performing studies in Latin American in cancer types different than breast and colorectal and a screening of other mutations in addition to the most popular mutations analyzed, such as c.1100delC.
F1000Research: Dataset 1: A complete list of mutations, before applying filters, in CHEK2 reported in the cBioPortal 10.5256/f1000research.9932.d14212948.
F1000Research: Dataset 2: A complete list of mutations, before applying filters, in CHEK2 reported in the ICGC 10.5256/f1000research.9932.d14213049.
F1000Research: Dataset 3: Mutations in CHEK2 identified in Latino American samples before applying filters (source:ExAC) 10.5256/f1000research.9932.d14213150.
F1000Research: Dataset 4: Variants reported in CHEK2 that have been associated with cancer according to data in the GWAS catalog. All of these variants were found in the cBioPortal or ICGC data 10.5256/f1000research.9932.d14213251.
F1000Research: Dataset 5: Number of individuals per cancer study and ethnicity in the TCGA cohort. Only studies in which at least one mutation in CHEK2 was found were included 10.5256/f1000research.9932.d14213352.
Conception and design of the work: CCL, GOS and RHAL. Data collection: RHAL and GOS. Data analysis: CCL, GOS and RHAL. Drafting of the article and critical revision: CCL, GOS, and RHAL. All authors were involved in the revision of the draft manuscript and have agreed to the final content.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: cancer genetics, molecular biology of carcinogenesis, epidemiology of cancer, pharmacogenetics
Competing Interests: No competing interests were disclosed.
References
1. Bell DW, Kim SH, Godwin AK, Schiripo TA, et al.: Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts.Int J Cancer. 2007; 121 (12): 2661-7 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 29 Nov 16 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)