Introduction
Platforms such as microarrays and, more recently, next generation sequencing have been leveraged to generate molecular profiles at the scale of entire systems. The global perspective gained using such approaches is potentially transformative. Transcriptome profiling enabled for instance the characterization of molecular perturbations that occur in the context of a wide range disease processes1–10. This in turn has provided opportunities for the discovery of biomarkers and for the development of novel therapeutic modalities3,11–13. More recently such systems-scale profiling of the blood transcriptome has also been used to monitor response to vaccines or therapeutic drugs14–19. The democratization of these approaches has led to proliferation of data in public repositories: over 1.7 million individual transcriptome profiles from more than 65,000 studies have been deposited to date in the NCBI Gene Expression Omnibus (GEO), a public repository of transcriptome profiles.
Taken together this vast body of “collective data” holds the promise of accelerating the pace of biomedical discovery by creating countless opportunities for identifying and filling critical knowledge gaps. Building tools that provide biomedical researchers with the ability to seamlessly interact with collections of datasets along with rich contextual information is essential in promoting insight and enabling knowledge discovery. To address this need we have developed an interactive data browsing and visualization web application, the Gene Expression Browser (GXB).
GXB was described in a recent publication and is available as open source software on GitHub20. This tool constitutes a simple interface for the browsing and interactive visualization of large volumes of heterogeneous data. Users can easily customize data plots by adding multiple layers of information, modifying the order of samples, and generating links that capture these settings, which can be inserted in email communications or in publications. Accessing the tool via these links also provides access to rich contextual information that is essential for data interpretation. This includes access to gene information and relevant literature, study design information, detailed sample information as well as ancillary data20.
In recent years, a large number of transcriptional studies have been conducted aiming at the characterization and functional classification of monocytes in health and disease. Monocytes are a population of immune cells found in the blood, bone marrow, and spleen. They constitute ~10% of the total circulating blood leukocytes in humans. They can remain in the blood circulation for up to 1–2 days, after which time, if they have not been recruited to a tissue, they die and are removed. They are considered the systemic reservoir of myeloid precursors for renewal of tissue macrophages and dendritic cells. Monocytes play a key role during immune response as professional phagocytes21,22, and producers of immune mediators23,24. Indeed, reports show that monocytes are recruited at the site of infections as innate effectors of the inflammatory response to microbes, killing pathogens via phagocytosis, production of reactive oxygen intermediate (ROIs)25, reactive nitrogen intermediate (RNIs)26,27, myeloperoxidase (MPO)28,29, and producing inflammatory cytokines30 that contribute to further amplifying the antimicrobial response31.
Human monocytes are derived from hematopoietic stem cells in the bone marrow and are released into peripheral blood circulation upon maturation. They are divided into three major subsets based on the expression of the cell surface markers CD14 and CD16. The most prevalent subset in the blood circulation, accounting for 90% of all monocytes, are the classical monocytes that express high levels of CD14 but low levels of CD16 (CD14++CD16-). The remaining 10% is divided into two subsets: intermediate monocytes with high expression of CD14 and CD16 (CD14++CD16+ or CD14+CD16+) and non-classical monocytes that express low levels of CD14 but high levels of CD16 (CD14dimCD16++ or CD14-CD16++)32–34. The factors that govern the migration of monocytes and roles that each subset plays during disease processes are not well understood. 1) In autoimmune diseases: Non-classical monocytes are regarded as crucial effectors in the pathogenesis of rheumatoid arthritis, ankylosing spondylitis35, systemic lupus erythematosus (SLE)36 and multiple sclerosis37. This monocyte subset carries a distinct inflammatory signature in patients with SLE36. Classical monocytes on the other hand have been shown to dominate the inflamed mucosa in Crohn’s disease38. Skewing of monocytes towards the intermediate subset has been observed in patients with autoimmune uveitis and linked to administration of glucocorticoid therapy39. 2) In cardiovascular diseases: circulating monocytes play a pivotal role by releasing cocktails of cytokines, factor and proteases that are involved in vascular growth40. Monocyte subsets show functional and phenotypic changes in cardiovascular diseases. The accumulation of classical monocytes is for instance a hallmark of progression of atherosclerosis41–43. An association between intermediate monocytes and cardiovascular events has also been documented with this monocyte subset being proportionally elevated following myocardial infarction or atrial fibrillation44,45 or in at risk subjects46. 3) In cancer: Intermediate monocytes are viewed as potential diagnostic indicators for colorectal cancer47. Another study has shown that elevated abundance of intermediate monocytes is associated with survival of adult or childhood acute lymphoblastic leukemia48. The changes of gene expression profiles in monocytes reveal high specificity for the tissue type and cancer histotype, and are induced in response to soluble factors released by the cancer cells in the primary or metastatic site49. Moreover, monocytes, comprising the monocyte-myeloid-derived suppressor cells population, from patients with metastatic breast cancer resemble the reprogrammed immunosuppressive monocytes in patients with severe infections, both by their surface and functional phenotype but also by their gene expression profile50. This signature of immunosuppression could therefore constitute a good biomarker for assessing disease progression. 4) In infections: monocytes are also key players in the immediate immune response to infectious agents as well as the subsequent development of the adaptive immune response51. Given the importance of classical and intermediate monocytes in pathogenesis of infectious and other inflammatory disorders, delineation of their functional and phenotypic characteristics has been studied extensively. The response mounted by classical monocytes has emerged as being critical for the control of a wide range of infectious diseases, including infections caused by bacteria52–57, parasites58 and fungi59. In contrast, intermediate monocytes have been associated with pathologic immune responses against bacteria60,61 and parasites62. In the context of HIV infection; CD14 expression is reduced on classical monocytes in chronically HIV-1 infected adults on anti-retroviral therapy63,64. Moreover, loss of CCR2 expressing non-classical monocytes is associated with cognitive impairment in antiretroviral therapy-naïve infected subjects65
. Altogether these findings indicate that monocytes are more than circulating precursors and have different effector functions in response to various infections and during inflammation. Clearly furthering our understanding of the role of monocyte subsets in health and disease will require many more studies, also we hope that the dataset compendium that we are making available to the research community via this publication can help support these endeavors.
In this data note we are making available via GXB a curated compendium of 93 public datasets relevant to human monocyte immunobiology, representing a total of 4,516 transcriptome profiles.
Materials and methods
Identification of monocyte datasets
Potentially relevant datasets deposited in GEO were identified using an advanced query based on the Bioconductor package GEOmetadb and the SQLite database that captures detailed information on the GEO data structure; https://www.bioconductor.org/packages/release/bioc/html/GEOmetadb.html66. The search query was designed to retrieve entries where the title and description contained the word Monocyte OR Monocytes, were generated from human samples, using Illumina or Affymetrix commercial platforms. The query result is appended with rich metadata from GEOmetadb that allows for manual filtering of the retrieved collection.
The relevance of each entry returned by this query was assessed individually. This process involved reading through the descriptions and examining the list of available samples and their annotations. Sometimes it was also necessary to review the original published report in which the design of the study and generation of the dataset is described in more detail. Using the search query, the results also returned a number of datasets that did not include profiles of monocytes but instead of “monocyte-derived dendritic cells” or “monocyte-derived macrophages”. During our manual screen these were excluded as were studies employing monocytic cell lines. Only studies including primary human monocyte profiles were retained. The datasets cover a broad range of studies investigating human monocyte immunobiology in the context of diseases and through comparison with diverse cell populations and study types as illustrated by a graphical representation of relative occurrences of terms in the descriptions of the studies loaded into our tool (Figure 1). A wide range of cell types and diseases are represented. Ultimately, the collection was comprised of 93 curated datasets. It includes datasets generated from studies profiling primary human CD14+ cells isolated from patients with autoimmune diseases (7), bacterial, virus and parasite infections (7), cancer (4), cardiovascular diseases (4), kidney diseases (4), as well as monocytes isolated from healthy subjects (58) (Figure 2). The 58 datasets in which monocytes were isolated from healthy subjects were classified based on whether profiling was conducted ex vivo or following in vitro experiments. In total 38 datasets were identified in which primary human CD14+ cells were stimulated or infected in in vitro experiments (Figure 2). Among the many noteworthy datasets, there are 8 datasets investigating differences between monocytes subsets; classical (CD14++CD16-), intermediate (CD14+CD16+) and non-classical monocytes (CD14-CD16++)32–34 [GXB: GSE16836, GSE18565, GSE25913, GSE34515, GSE35457, GSE51997, GSE60601, GSE66936]. Another dataset from Banchereau and colleagues investigated responses of monocyte and dendritic cells to 13 different vaccines in vitro67 [GXB: GSE44721]. The datasets that comprise our collection are listed in Table 1 and can be browsed interactively in GXB.

Figure 1. Thematic composition of the dataset collection.
Word frequencies extracted from text descriptions of the studies loaded into the GXB tool are depicted as a word cloud. The size of the words is proportional to their frequency.
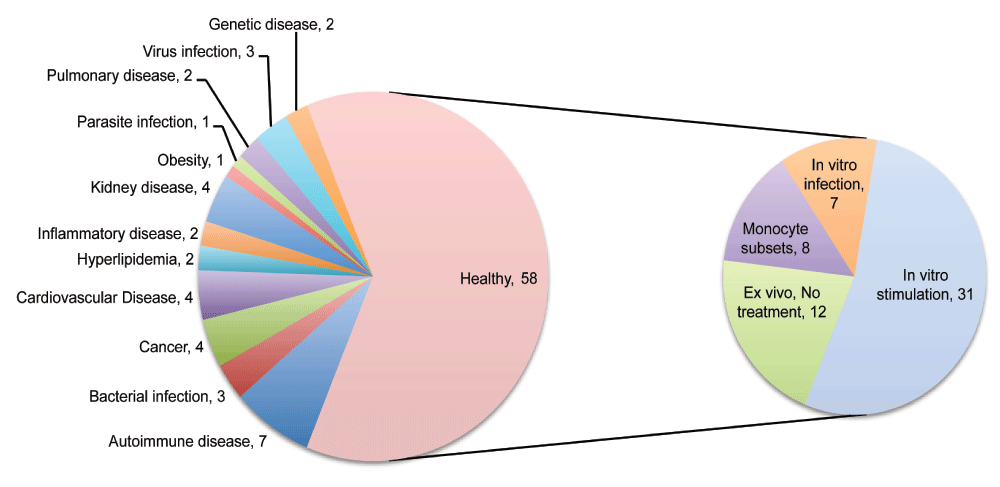
Figure 2. Break down of the dataset collection by category.
The pie chart on the left panel indicates dataset frequencies by disease status. The chart on the right panel indicates the type of studies carried out for the 58 datasets consisting of monocyte obtained exclusively from healthy donors.
Table 1. List of datasets constituting the collection.
| Title | Platforms | Diseases | Number
of
samples | Experiments | GEO ID | Ref |
|---|
Interaction of bone marrow stroma and monocytes: bone marrow stromal
cell lines cultured with monocytes
| Affymetrix | Healthy | 8 |
In vitro
|
GSE10595
|
68
|
Monocyte gene expression profiling in familial combined hyperlipidemia and
its modification by atorvastatin treatment
| Affymetrix | Familial combined
hyperlipidemia | 9 |
In vitro
|
GSE11393
|
69
|
|
Performance comparison of Affymetrix and Illumina microarray technologies
| Affymetrix | Acute coronary syndrome | 10 |
Ex vivo
|
GSE11430
|
70
|
Gene expression profiling in pediatric meningococcal sepsis reveals
dynamic changes in NK-cell and cytotoxic molecules
| Affymetrix | Meningococcal sepsis | 41 |
Ex vivo
|
GSE11755
| N/A |
Effect of interferon-gamma on macrophage differentiation and response to
Toll-like receptor ligands
| Affymetrix | Healthy | 10 |
In vitro
|
GSE11864
|
71
|
|
Human monocyte and dendritic Cell Subtype Gene Arrays
| Affymetrix | Healthy | 8 |
Ex vivo
|
GSE11943
|
72
|
|
Microarray analysis of human monocytes infected with Francisella tularensis
| Affymetrix | Healthy | 14 |
In vitro
|
GSE12108
|
73
|
|
Human blood monocyte profile in Ventilator-Associated Pneumonia patients
| Affymetrix | Pneumonia | 60 |
Ex vivo
|
GSE12838
| N/A |
|
Quercetin supplementation and CD14+ monocyte gene expression
| Affymetrix | Healthy | 6 |
Ex vivo
|
GSE13899
|
74
|
|
Effects of PMN-Ectosomes on human macrophages
| Affymetrix | Healthy | 16 |
In vitro
|
GSE14419
| N/A |
Homogeneous monocytes and macrophages from hES cells following
coculture-free differentiation in M-CSF and IL-3
| Affymetrix | Healthy | 9 |
Ex vivo
|
GSE15791
|
75
|
|
Expression data from human macrophages
| Affymetrix | Healthy | 38 |
In vitro
|
GSE16385
|
76
|
Transcriptional profiling of CD16+ and CD16- peripheral blood monocytes
from healthy individuals
| Affymetrix | Healthy | 8 |
Ex vivo
|
GSE16836
|
32
|
COPD-Specific Gene Expression Signatures of Alveolar Macrophages
as well as Peripheral Blood Monocytes Overlap and Correlate with Lung
Function
| Affymetrix | Chronic Obstructive
Pulmonary Disease | 12 |
Ex vivo
|
GSE16972
|
77
|
Loss-of-function mutations in REP-1 affect intracellular vesicle transport in
fibroblasts and monocytes of CHM patients
| Affymetrix | Choroideremia | 15 |
Ex vivo
|
GSE17549
|
78
|
Effect of two weeks erythropoietin treatment on monocyte transcriptomes of
cardiorenal patients
| Illumina | Cardiorenal syndrome | 48 |
Ex vivo
|
GSE17582
| N/A |
|
Comparison of gene expression profiles between human monocyte subsets
| Affymetrix | Healthy | 6 |
Ex vivo
|
GSE18565
|
79
|
Subpopulations of CD163 positive macrophages are classically activated in
psoriasis
| Illumina | Psoriasis | 58 |
Ex vivo
|
GSE18686
|
80
|
Mycobacterium tuberculosis Chaperonin 60.1 has Bipolar Effects on Human
peripheral blood-derived Monocytes
| Affymetrix | Healthy | 21 |
In vitro
|
GSE18794
| N/A |
|
Blood Transcriptional Profiles of Active TB (Separated cell)
| Illumina | Tuberculosis | 44 |
Ex vivo
|
GSE19443
|
11
|
|
Filaria induced monocyte dysfunction and its reversal following treatment
| Affymetrix | Filariasis | 14 |
Ex vivo
|
GSE2135
|
81
|
Ubiquinol-induced gene expression signatures are translated into reduced
erythropoiesis and LDL cholesterol levels in humans
| Affymetrix | Healthy | 6 |
Ex vivo
|
GSE21351
| 82 |
|
Monocyte vs Macrophage Study
| Affymetrix | Healthy | 6 |
In vitro
|
GSE22373
|
83
|
Monocyte gene expression patterns distinguish subjects with and without
atherosclerosis
| Illumina | Carotid atherosclerosis | 95 |
Ex vivo
|
GSE23746
| N/A |
Deconvoluting Early Post-Transplant Immunity Using Purified Cell Subsets
Reveals Functional Networks Not Evident by Whole Blood Analysis
| Affymetrix | Kidney Transplantation | 179 |
Ex vivo
|
GSE24223
|
84
|
Cooperative and redundant signaling of leukotriene B4 and leukotriene D4
in human monocytes
| Affymetrix | Healthy | 10 |
In vitro
|
GSE24869
|
85
|
Gene expression profiling of the classical (CD14++CD16-), intermediate
(CD14++CD16+) and nonclassical (CD14+CD16+) human monocyte
subsets
| Illumina | Healthy | 24 |
Ex vivo
|
GSE25913
|
34
|
Direct Cell Conversion of Human Fibroblasts to Monocytic phagocytes by
Forced Expression of Monocytic Regulatory Network Elements
| Illumina | Dermatomyositis | 15 |
Ex vivo
|
GSE27304
| N/A |
|
cMyb and vMyb in human monocytes
| Affymetrix | Healthy | 6 |
In vitro
|
GSE2816
|
86
|
Temporal transcriptional changes in human monocytes following acute
myocardial infarction: The GerMIFs monocyte expression study
| Illumina | Acute myocardial infarction | 76 |
Ex vivo
|
GSE28454
| N/A |
|
mRNA expression profiling of human immune cell subset (Roche)
| Affymetrix | Healthy | 47 |
Ex vivo
|
GSE28490
|
87
|
|
mRNA expression profiling of human immune cell subsets (HUG)
| Affymetrix | Healthy | 33 |
Ex vivo
|
GSE28491
|
87
|
Changes in gene expression profiles in patients with 5q- syndrome in
CD14+ monocytes caused by lenalidomide treatment
| Illumina | 5q- syndrome | 17 |
Ex vivo
|
GSE31460
| N/A |
Genome-wide analysis of lupus immune complex stimulation of purified
CD14+ monocytes and how this response is regulated by C1q
| Illumina | Healthy | 8 |
In vitro
|
GSE32278
|
88
|
Transcriptome analysis of circulating monocytes in obese patients before
and three months after bariatric surgery
| Illumina | Obesity | 48 |
Ex vivo
|
GSE32575
|
89
|
|
CD4 on human monocytes
| Affymetrix | Healthy | 6 |
In vitro
|
GSE32939
|
90
|
Peripheral Blood Monocyte Gene Expression in Recent-Onset Type 1
Diabetes
| Illumina | Type 1 Diabetes | 22 |
Ex vivo
|
GSE33440
|
91
|
Traffic-related Particulate Matter Upregulates Allergic Responses by a
Notch-pathway Dependent Mechanism
| Affymetrix | Healthy | 16 |
In vitro
|
GSE34025
| N/A |
|
Human monocyte activation with NOD2L vs. TLR2/1L
| Affymetrix | Healthy | 45 |
In vitro
|
GSE34156
|
92
|
Bacillus anthracis' lethal toxin induces broad transcriptional responses in
human peripheral monocyte
| Affymetrix | Healthy | 8 |
In vitro
|
GSE34407
|
93
|
Gene expression profiles of human blood classical monocytes
(CD14++CD16-), CD16 positive monocytes (CD14+16++ and
CD14++CD16+), and CD1c+ CD19- dendritic cells
| Affymetrix | Healthy | 9 |
Ex vivo
|
GSE34515
| N/A |
Genome-wide analysis of monocytes and T cells' response to interferon
beta
| Illumina | Healthy | 12 |
In vitro
|
GSE34627
|
94
|
Highly pathogenic influenza virus inhibit Inflammatory Responses in
Monocytes via Activation of the Rar-Related Orphan Receptor Alpha
(RORalpa)
| Affymetrix | Healthy | 12 |
In vitro
|
GSE35283
| N/A |
|
Transcriptome profiles of human monocyte and dendritic cell subsets
| Illumina | Healthy | 49 |
Ex vivo
|
GSE35457
|
95
|
|
Influenza virus A infected monocytes
| Illumina | Healthy | 6 |
In vitro
|
GSE35473
|
96
|
|
PGE2-induced OSM expression
| Affymetrix | Chronic wound | 6 |
Ex vivo
|
GSE36995
|
97
|
Inflammatory Expression Profiles in Monocyte to Macrophage Differentiation
amongst Patients with Systemic Lupus Erythematosus and Healthy Controls
with and without an Atherosclerosis Phenotype
| Illumina | Systemic lupus
erythematosus | 72 |
Ex vivo
|
GSE37356
| N/A |
New insights into key genes and pathways involved in the pathogenesis of
HLA-B27-associated acute anterior uveitis
| Affymetrix | Acute anterior uveitis | 6 |
In vitro
|
GSE37588
| N/A |
|
Analysis of blood myelomonocytic cells from RCC patients
| Illumina | Renal cell carcinoma | 8 |
Ex vivo
|
GSE38424
|
98
|
|
Nanotoxicogenomic study of ZnO and TiO2 responses
| Illumina | Healthy | 90 |
In vitro
|
GSE39316
| N/A |
Macrophage Microvesicles Induce Macrophage Differentiation and miR-223
Transfer
| Affymetrix | Healthy | 24 |
In vitro
|
GSE41889
|
99
|
|
TREM-1 is a novel therapeutic target in Psoriasis
| Affymetrix | Psoriasis | 15 |
In vitro
|
GSE42305
|
100
|
|
Comparison study between Uremic patient with Healthy control
| Affymetrix | Chronic kidney disease | 6 |
Ex vivo
|
GSE43484
| N/A |
Microarray analysis of IL-10 stimulated adherent peripheral blood
mononuclear cells
| Affymetrix | Healthy | 8 |
In vitro
|
GSE43700
|
101
|
|
Monocytes and Dendritic cells stimulated by 13 human vaccines and LPS
| Illumina | Vaccination | 128 |
In vitro
|
GSE44721
|
67
|
Gene expression profile of human monocytes stimulated with all-trans
retinoic acid (ATRA) or 1,25a-dihydroxyvitamin D3 (1,25D3)
| Affymetrix | Healthy | 12 |
In vitro
|
GSE46268
|
102
|
|
Transcriptome analysis of blood monocytes from sepsis patients
| Illumina | Sepsis | 44 |
Ex vivo
|
GSE46955
|
103
|
Tumor-educated circulating monocytes are powerful specific biomarkers for
diagnosis of colorectal cancer
| Illumina | Colorectal Cancer | 93 |
Ex vivo
|
GSE47756
|
49
|
|
Similarities and differences between macrophage polarized gene profiles
| Illumina | Healthy | 12 |
In vitro
|
GSE49240
|
104
|
|
The effect of cell subset isolation method on gene expression in leukocytes.
| Illumina | Healthy | 50 |
Ex vivo
|
GSE50008
| N/A |
|
Transcriptome analysis of HIV-infected peripheral blood monocytes
| Illumina | HIV | 86 |
Ex vivo
|
GSE50011
|
105
|
Gene expression profiles in T-lymphocytes and Monocytes of participants of
the Tour de France 2005
| Affymetrix | Healthy | 66 |
Ex vivo
|
GSE5105
| N/A |
|
Effects of exercise on gene expression level in human monocytes
| Affymetrix | Healthy | 24 |
Ex vivo
|
GSE51835
|
106
|
T helper lymphocyte- and monocyte-specific type I interferon (IFN)
signatures in autoimmunity and viral infection.
| Affymetrix | Autoimmune diseases | 36 |
Ex vivo
|
GSE51997
|
107
|
Longitudinal comparison of monocytes from an HIV viremic vs avirmeic
state
| Affymetrix | HIV | 16 |
Ex vivo
|
GSE5220
|
108
|
|
Expression data from monocytes and monocyte derived macrophages
| Affymetrix | Healthy | 12 |
In vitro
|
GSE52647
| N/A |
Transcriptome analysis of primary monocytes from HIV+ patients with
differential responses to therapy
| Illumina | HIV | 14 |
Ex vivo
|
GSE52900
|
109
|
|
Human blood monocyte response to IL-17A in culture
| Affymetrix | Healthy | 6 |
In vitro
|
GSE54884
| N/A |
Divergent genome wide transcriptional profiles from immune cell subsets
isolated from SLE patients with different ancestral backgrounds
| Illumina | Systemic lupus
erythematosus | 208 |
Ex vivo
|
GSE55447
|
110
|
Cell Specific Expression & Pathway Analyses Reveal Novel Alterations in
Trauma-Related Human T-Cell & Monocyte Pathways
| Affymetrix | Trauma patients | 42 |
Ex vivo
|
GSE5580
|
111
|
|
Immune Variation Project (ImmVar) [CD14]
| Affymetrix | Healthy | 485 |
Ex vivo
|
GSE56034
| N/A |
|
Transcriptomics of human monocytes
| Illumina | Healthy | 1202 |
Ex vivo
|
GSE56045
|
112
|
|
Effect of vitamin D treatment on human monocyte
| Affymetrix | Healthy | 16 |
In vitro
|
GSE56490
| NA |
Monocytes of patients with familial hypercholesterolemia show alterations in
cholesterol metabolism
| Affymetrix | Hypercholesterolemia | 23 |
Ex vivo
|
GSE6054
|
113
|
Gene expression data from CD14++ CD16- classical monocytes from
healthy volunteers and patients with pancreatic ductal adenocarcinoma
| Affymetrix | Pancreatic ductal
adenocarcinoma | 12 |
Ex vivo
|
GSE60601
| N/A |
|
Activation of the JAK/STAT pathway in Behcet's Disease
| Affymetrix | Behcet’s Disease | 29 |
Ex vivo
|
GSE61399
| N/A |
Alarmins MRP8 and MRP14 induce stress-tolerance in phagocytes under
sterile inflammatory conditions
| Illumina | Sterile Inflammation | 12 |
In vitro
|
GSE61477
| N/A |
|
GM-CSF induced gene-regulation in human monocytes
| Affymetrix | Healthy | 6 |
In vitro
|
GSE63662
|
114
|
|
Treatment of human monocytes with TLR7 or TLR8 agonists
| Affymetrix | Healthy | 9 |
In vitro
|
GSE64480
|
115
|
Restricted Dendritic Cell and Monocyte Progenitors in Human Cord Blood
and Bone Marrow
| Illumina | Healthy | 36 |
Ex vivo
|
GSE65128
|
116
|
Interleukin-1- and Type I Interferon-Dependent Enhanced Immunogenicity
of an NYVAC-HIV-1 Env-Gag-Pol-Nef Vaccine Vector with Dual Deletions of
Type I and Type II Interferon-Binding Proteins
| Illumina | Vaccination | 20 |
In vitro
|
GSE65412
| NA |
Comparative analysis of monocytes from healthy donors, patients with
metastatic breast cancer, sepsis or tuberculosis.
| Illumina | Breast cancer and Bacterial
infection | 13 |
Ex vivo
|
GSE65517
|
50
|
Expression data from intermediate monocytes from healthy donors and
autoimmune uveitis patients
| Affymetrix | Autoimmune uveitis | 21 |
Ex vivo
|
GSE66936
|
39
|
Induction of Dendritic Cell-like Phenotype in Macrophages during Foam Cell
Formation
| Affymetrix | Healthy | 22 |
In vitro
|
GSE7138
|
117
|
Genome Wide Gene Expression Study of Circulating Monocytes in human
with extremely high vs. low bone mass
| Affymetrix | Healthy | 26 |
Ex vivo
|
GSE7158
| N/A |
Genomic profiles for human peripheral blood T cells, B cells, natural killer
cells, monocytes, and polymorphonuclear cells: comparisons to ischemic
stroke, migraine, and Tourette syndrome
| Affymetrix | Healthy | 18 |
Ex vivo
|
GSE72642
|
118
|
Expression data from monocytes of individuals with different collateral flow
index CFI
| Affymetrix | Coronary artery disease | 160 |
Ex vivo
|
GSE7638
|
39
|
Leukotriene D4 induces gene expression in human monocytes through
cysteinyl leukotriene type I receptor
| Affymetrix | Healthy | 8 |
In vitro
|
GSE7807
|
119
|
|
Gene expression profile during monocytes to macrophage differentiation
| Affymetrix | Healthy | 9 |
In vitro
|
GSE8286
| N/A |
Toll-like receptor triggering of a vitamin D-mediated human antimicrobial
response
| Affymetrix | Healthy | 50 |
In vitro
|
GSE8921
|
120
|
|
TRAIL Is a Novel Antiviral Protein against Dengue Virus
| Affymetrix | Dengue | 10 |
In vitro
|
GSE9378
| NA |
Gene Expression-Based High Throughput Screening: APL Treatment with
Candidate Compounds
| Affymetrix | Leukemia | 24 |
Ex vivo
|
GSE976
|
121
|
|
Innate immune responses to TREM-1 activation
| Affymetrix | Healthy | 11 |
In vitro
|
GSE9988
|
122
|
Dataset upload and annotation on GXB
Once a final selection was made each dataset was downloaded from GEO in the SOFT file format. It was in turn uploaded on an instance of the Gene Expression Browser (GXB) hosted on the Amazon Web Services cloud. Available sample and study information were also uploaded. Samples were grouped according to possible interpretations of study results and ranking based on the different group comparisons that were computed (e.g. comparing monocyte isolated from case vs controls in studies where profiling was performed ex-vivo; or stimulated vs medium control in in vitro experiments).
Short Gene Expression Brower tutorial
The GXB software has been described in detail in a recent publication20. This custom software interface provides users with a means to easily navigate and filter the dataset collection available at http://monocyte.gxbsidra.org/dm3/landing.gsp. A web tutorial is also available online: http://monocyte.gxbsidra.org/dm3/tutorials.gsp#gxbtut. Briefly, datasets of interest can be quickly identified either by filtering using criteria from pre-defined lists on the left or by entering a query term in the search box at the top of the dataset navigation page. Clicking on one of the studies listed in the dataset navigation page opens a viewer designed to provide interactive browsing and graphic representations of large-scale data in an interpretable format. This interface is designed to present ranked gene lists and display expression results graphically in a context-rich environment. Selecting a gene from the rank ordered list on the left of the data-viewing interface will display its expression values graphically in the screen’s central panel. Directly above the graphical display drop down menus give users the ability: a) To change how the gene list is ranked; this allows the user to change the method used to rank the genes, or to include only genes that are selected for specific biological interest; b) To change sample grouping (Group Set button), in some datasets a user can switch between groups based on cell type to groups based on disease type, for example; c) To sort individual samples within a group based on associated categorical or continuous variables (e.g. gender or age); d) To toggle between the bar chart view and a box plot view, with expression values represented as a single point for each sample. Samples are split into the same groups whether displayed as a bar chart or box plot; e) To provide a color legend for the sample groups; f) To select categorical information that is to be overlaid at the bottom of the graph. For example, the user can display gender or treatment status in this manner; g) To provide a color legend for the categorical information overlaid at the bottom of the graph; and h) To download the graph as a png image or csv file for performing a separate analysis. Measurements have no intrinsic utility in absence of contextual information. It is this contextual information that makes the results of a study or experiment interpretable. It is therefore important to capture, integrate and display information that will give users the ability to interpret data and gain new insights from it. We have organized this information under different tabs directly above the graphical display. The tabs can be hidden to make more room for displaying the data plots, or revealed by clicking on the blue “show info panel” button on the top right corner of the display. Information about the gene selected from the list on the left side of the display is available under the “Gene” tab. Information about the study is available under the “Study” tab. Information available about individual samples is provided under the “Sample” tab. Rolling the mouse cursor over a bar chart's element while displaying the “Sample” tab lists any clinical, demographic, or laboratory information available for the selected sample. Finally, the “Downloads” tab allows advanced users to retrieve the original dataset for analysis outside this tool. It also provides all available sample annotation data for use alongside the expression data in third party analysis software. Other functionalities are provided under the “Tools” drop-down menu located in the top right corner of the user interface. Some of the notable functionalities available through this menu include: a) Annotations, which provides access to all the ancillary information about the study, samples and dataset organized across different tabs; b) Cross-project view, which provides the ability for a given gene to browse through all available studies; c) Copy link, which generates a mini-URL encapsulating information about the display settings in use and that can be saved and shared with others (clicking on the envelope icon on the toolbar inserts the url in an email message via the local email client); and d) Chart options, which gives user the option to customize chart labels.
Dataset validation
Quality control checks were performed with the examination of profiles of relevant biological indicators. Known leukocyte markers were used, such as CD14, which is expressed by monocytes and macrophages; as well as markers that would indicate significant contamination of the sample by other leukocyte populations: such as CD3, a T-cells marker; CD19, a B-cell marker; CD56, an NK cell marker (Figure 3; The expression of the CD14 marker across all studies can be checked using the cross project functionality of GXB: http://monocyte.gxbsidra.org/dm3/geneBrowser/crossProject?probeID=201743_at&geneSymbol=CD14&geneID=929). We have systematically verified that expression of the genes encoding those surface markers was consistent with grouping labels provided by depositors. In addition, expression of the XIST transcripts, in which expression is gender-specific, was also examined to determine its concordance with demographic information provided with the GEO submission (expression of XIST should be high in females and low in males).

Figure 3. Illustrative example showing the abundance levels of CD14 transcripts across samples in a given study.
The expression of this gene is indicative of the purity of primary human monocyte preparation; this marker is expected to be high in monocyte preparations and low in other leukocyte populations. In this view of the GXB expression of CD14 can be visualized across projects listed on the left.
Data availability
All datasets included in our curated collection are also available publically via the NCBI GEO website: http://www.ncbi.nlm.nih.gov/geo/; and are referenced throughout the manuscript by their GEO accession numbers (e.g. GSE25913). Signal files and sample description files can also be downloaded from the GXB tool under the “downloads” tab.
Author contributions
DR: curated, uploaded and annotated datasets, and drafted the manuscript. SB: installed the software, uploaded datasets, programmed portions of the web application, and tested the software, and assisted in drafting the manuscript. SP: participated in the design of the software, programmed portions of the original web application, installed the software, and tested the software, and assisted in drafting the manuscript. CQ: participated in designed and programmed portions of the original web application, tested the software, and assisted in drafting the manuscript. DC: participated in software design, tested the software, and drafted the manuscript.
Competing interests
No competing interests were disclosed.
Grant information
DR, SB and DC received support from the Qatar Foundation.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgement
The authors would like to acknowledge all the investigators who decided to make their datasets publically available by sharing them in GEO.
Faculty Opinions recommendedReferences
- 1.
Bennett L, Palucka AK, Arce E, et al.:
Interferon and granulopoiesis signatures in systemic lupus erythematosus blood.
J Exp Med.
2003; 197(6): 711–23. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 2.
Griffiths MJ, Shafi MJ, Popper SJ, et al.:
Genomewide analysis of the host response to malaria in Kenyan children.
J Infect Dis.
2005; 191(10): 1599–611. PubMed Abstract
| Publisher Full Text
- 3.
Mejias A, Dimo B, Suarez NM, et al.:
Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection.
PLoS Med.
2013; 10(11): e1001549. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 4.
Moal V, Textoris J, Ben Amara A, et al.:
Chronic hepatitis E virus infection is specifically associated with an interferon-related transcriptional program.
J Infect Dis.
2013; 207(1): 125–32. PubMed Abstract
| Publisher Full Text
- 5.
Mostafavi S, Battle A, Zhu X, et al.:
Type I interferon signaling genes in recurrent major depression: increased expression detected by whole-blood RNA sequencing.
Mol Psychiatry.
2014; 19(12): 1267–74. PubMed Abstract
| Publisher Full Text
- 6.
Novershtern N, Subramanian A, Lawton LN, et al.:
Densely interconnected transcriptional circuits control cell states in human hematopoiesis.
Cell.
2011; 144(2): 296–309. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 7.
Panelli MC, Wang E, Phan G, et al.:
Gene-expression profiling of the response of peripheral blood mononuclear cells and melanoma metastases to systemic IL-2 administration.
Genome Biol.
2002; 3(7): RESEARCH0035. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 8.
Pascual V, Allantaz F, Patel P, et al.:
How the study of children with rheumatic diseases identified interferon-alpha and interleukin-1 as novel therapeutic targets.
Immunol Rev.
2008; 223(1): 39–59. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 9.
Smih F, Desmoulin F, Berry M, et al.:
Blood signature of pre-heart failure: a microarrays study.
PLoS One.
2011; 6(6): e20414. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 10.
Stamova B, Xu H, Jickling G, et al.:
Gene expression profiling of blood for the prediction of ischemic stroke.
Stroke.
2010; 41(10): 2171–7. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 11.
Berry MP, Graham CM, McNab FW, et al.:
An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis.
Nature.
2010; 466(7309): 973–7. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 12.
Martínez-Llordella M, Lozano JJ, Puig-Pey I, et al.:
Using transcriptional profiling to develop a diagnostic test of operational tolerance in liver transplant recipients.
J Clin Invest.
2008; 118(8): 2845–57. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 13.
Newell KA, Asare A, Kirk AD, et al.:
Identification of a B cell signature associated with renal transplant tolerance in humans.
J Clin Invest.
2010; 120(6): 1836–47. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 14.
Gaucher D, Therrien R, Kettaf N, et al.:
Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses.
J Exp Med.
2008; 205(13): 3119–31. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 15.
Hecker M, Hartmann C, Kandulski O, et al.:
Interferon-beta therapy in multiple sclerosis: the short-term and long-term effects on the patients' individual gene expression in peripheral blood.
Mol Neurobiol.
2013; 48(3): 737–56. PubMed Abstract
| Publisher Full Text
- 16.
Li S, Rouphael N, Duraisingham S, et al.:
Molecular signatures of antibody responses derived from a systems biology study of five human vaccines.
Nat Immunol.
2014; 15(2): 195–204. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 17.
Obermoser G, Presnell S, Domico K, et al.:
Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines.
Immunity.
2013; 38(4): 831–44. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 18.
Oswald M, Curran ME, Lamberth SL, et al.:
Modular analysis of peripheral blood gene expression in rheumatoid arthritis captures reproducible gene expression changes in tumor necrosis factor responders.
Arthritis Rheumatol.
2015; 67(2): 344–51. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 19.
Querec TD, Akondy RS, Lee EK, et al.:
Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans.
Nat Immunol.
2009; 10(1): 116–25. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 20.
Speake C, Presnell S, Domico K, et al.:
An interactive web application for the dissemination of human systems immunology data.
J Transl Med.
2015; 13: 196. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 21.
Auffray C, Sieweke MH, Geissmann F:
Blood monocytes: development, heterogeneity, and relationship with dendritic cells.
Annu Rev Immunol.
2009; 27: 669–92. PubMed Abstract
| Publisher Full Text
- 22.
van Furth R, Cohn ZA, Hirsch JG, et al.:
The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells.
Bull World Health Organ.
1972; 46(6): 845–52. PubMed Abstract
| Free Full Text
- 23.
Narni-Mancinelli E, Soudja SM, Crozat K, et al.:
Inflammatory monocytes and neutrophils are licensed to kill during memory responses in vivo.
PLoS Pathog.
2011; 7(12): e1002457. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 24.
Grimm MJ, Vethanayagam RR, Almyroudis NG, et al.:
Monocyte- and macrophage-targeted NADPH oxidase mediates antifungal host defense and regulation of acute inflammation in mice.
J Immunol.
2013; 190(8): 4175–84. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 25.
Fang FC:
Antimicrobial reactive oxygen and nitrogen species: concepts and controversies.
Nat Rev Microbiol.
2004; 2(10): 820–32. PubMed Abstract
| Publisher Full Text
- 26.
Dinauer MC, Deck MB, Unanue ER:
Mice lacking reduced nicotinamide adenine dinucleotide phosphate oxidase activity show increased susceptibility to early infection with Listeria monocytogenes.
J Immunol.
1997; 158(12): 5581–3. PubMed Abstract
- 27.
Endres R, Luz A, Schulze H, et al.:
Listeriosis in p47phox-/- and TRp55-/- mice: protection despite absence of ROI and susceptibility despite presence of RNI.
Immunity.
1997; 7(3): 419–32. PubMed Abstract
| Publisher Full Text
- 28.
Aratani Y, Koyama H, Nyui S, et al.:
Severe impairment in early host defense against Candida albicans in mice deficient in myeloperoxidase.
Infect Immun.
1999; 67(4): 1828–36. PubMed Abstract
| Free Full Text
- 29.
Albrecht D, Jungi TW:
Luminol-enhanced chemiluminescence induced in peripheral blood-derived human phagocytes: obligatory requirement of myeloperoxidase exocytosis by monocytes.
J Leukoc Biol.
1993; 54(4): 300–6. PubMed Abstract
- 30.
Gavrilin MA, Bouakl IJ, Knatz NL, et al.:
Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release.
Proc Natl Acad Sci U S A.
2006; 103(1): 141–6. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 31.
Serbina NV, Jia T, Hohl TM, et al.:
Monocyte-mediated defense against microbial pathogens.
Annu Rev Immunol.
2008; 26: 421–52. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 32.
Ancuta P, Liu KY, Misra V, et al.:
Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16- monocyte subsets.
BMC Genomics.
2009; 10: 403. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 33.
Frankenberger M, Hofer TP, Marei A, et al.:
Transcript profiling of CD16-positive monocytes reveals a unique molecular fingerprint.
Eur J Immunol.
2012; 42(4): 957–74. PubMed Abstract
| Publisher Full Text
- 34.
Wong KL, Tai JJ, Wong WC, et al.:
Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets.
Blood.
2011; 118(5): e16–31. PubMed Abstract
| Publisher Full Text
- 35.
Aeberli D, Kamgang R, Balani D, et al.:
Regulation of peripheral classical and non-classical monocytes on infliximab treatment in patients with rheumatoid arthritis and ankylosing spondylitis.
RMD Open.
2016; 2(1): e000079. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 36.
Mukherjee R, Kanti Barman P, Kumar Thatoi P, et al.:
Non-Classical monocytes display inflammatory features: Validation in Sepsis and Systemic Lupus Erythematous.
Sci Rep.
2015; 5: 13886. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 37.
Kooij G, Braster R, Koning JJ, et al.:
Trichuris suis induces human non-classical patrolling monocytes via the mannose receptor and PKC: implications for multiple sclerosis.
Acta Neuropathol Commun.
2015; 3: 45. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 38.
Korkosz M, Bukowska-Strakova K, Sadis S, et al.:
Monoclonal antibodies against macrophage colony-stimulating factor diminish the number of circulating intermediate and nonclassical (CD14++CD16+/CD14+CD16++) monocytes in rheumatoid arthritis patient.
Blood.
2012; 119(22): 5329–30. PubMed Abstract
| Publisher Full Text
- 39.
Liu B, Dhanda A, Hirani S, et al.:
CD14++CD16+ Monocytes Are Enriched by Glucocorticoid Treatment and Are Functionally Attenuated in Driving Effector T Cell Responses.
J Immunol.
2015; 194(11): 5150–60. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 40.
Heil M, Eitenmüller I, Schmitz-Rixen T, et al.:
Arteriogenesis versus angiogenesis: similarities and differences.
J Cell Mol Med.
2006; 10(1): 45–55. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 41.
Drechsler M, Soehnlein O:
The complexity of arterial classical monocyte recruitment.
J Innate Immun.
2013; 5(4): 358–66. PubMed Abstract
| Publisher Full Text
- 42.
Sala F, Cutuli L, Grigore L, et al.:
Prevalence of classical CD14++/CD16- but not of intermediate CD14++/CD16+ monocytes in hypoalphalipoproteinemia.
Int J Cardiol.
2013; 168(3): 2886–9. PubMed Abstract
| Publisher Full Text
- 43.
Williams H, Cassorla G, Pertsoulis N, et al.:
Human classical monocytes display unbalanced M1/M2 phenotype with increased atherosclerotic risk and presence of disease.
Int Angiol.
2016. PubMed Abstract
- 44.
Lu W, Zhang Z, Fu C, et al.:
Intermediate monocytes lead to enhanced myocardial remodelling in STEMI patients with diabetes.
Int Heart J.
2015; 56(1): 22–8. PubMed Abstract
| Publisher Full Text
- 45.
Suzuki A, Fukuzawa K, Yamashita T, et al.:
Circulating intermediate CD14++CD16+monocytes are increased in patients with atrial fibrillation and reflect the functional remodelling of the left atrium.
Europace.
2016; pii: euv422. PubMed Abstract
| Publisher Full Text
- 46.
Rogacev KS, Zawada AM, Emrich I, et al.:
Lower Apo A-I and lower HDL-C levels are associated with higher intermediate CD14++CD16+ monocyte counts that predict cardiovascular events in chronic kidney disease.
Arterioscler Thromb Vasc Biol.
2014; 34(9): 2120–7. PubMed Abstract
| Publisher Full Text
- 47.
Schauer D, Starlinger P, Reiter C, et al.:
Intermediate monocytes but not TIE2-expressing monocytes are a sensitive diagnostic indicator for colorectal cancer.
PLoS One.
2012; 7(9): e44450. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 48.
Sulicka J, Surdacki A, Mikolajczyk T, et al.:
Elevated markers of inflammation and endothelial activation and increased counts of intermediate monocytes in adult survivors of childhood acute lymphoblastic leukemia.
Immunobiology.
2013; 218(5): 810–6. PubMed Abstract
| Publisher Full Text
- 49.
Hamm A, Prenen H, Van Delm W, et al.:
Tumour-educated circulating monocytes are powerful candidate biomarkers for diagnosis and disease follow-up of colorectal cancer.
Gut.
2015; pii: gutjnl-2014-308988. PubMed Abstract
| Publisher Full Text
- 50.
Bergenfelz C, Larsson AM, von Stedingk K, et al.:
Systemic Monocytic-MDSCs Are Generated from Monocytes and Correlate with Disease Progression in Breast Cancer Patients.
PLoS One.
2015; 10(5): e0127028. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 51.
Hume DA:
The mononuclear phagocyte system.
Curr Opin Immunol.
2006; 18(1): 49–53. PubMed Abstract
| Publisher Full Text
- 52.
Kolseth IB, Agren J, Sundvold-Gjerstad V, et al.:
9-cis retinoic acid inhibits inflammatory responses of adherent monocytes and increases their ability to induce classical monocyte migration.
J Innate Immun.
2012; 4(2): 176–86. PubMed Abstract
| Publisher Full Text
- 53.
Jakubzick C, Gautier EL, Gibbings SL, et al.:
Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes.
Immunity.
2013; 39(3): 599–610. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 54.
Fallows D, Peixoto B, Kaplan G, et al.:
Mycobacterium leprae alters classical activation of human monocytes in vitro.
J Inflamm (Lond).
2016; 13: 8. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 55.
Wantha S, Alard JE, Megens RT, et al.:
Neutrophil-derived cathelicidin promotes adhesion of classical monocytes.
Circ Res.
2013; 112(5): 792–801. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 56.
Shibata T, Takemura N, Motoi Y, et al.:
PRAT4A-dependent expression of cell surface TLR5 on neutrophils, classical monocytes and dendritic cells.
Int Immunol.
2012; 24(10): 613–23. PubMed Abstract
| Publisher Full Text
- 57.
Anbazhagan K, Duroux-Richard I, Jorgensen C, et al.:
Transcriptomic network support distinct roles of classical and non-classical monocytes in human.
Int Rev Immunol.
2014; 33(6): 470–89. PubMed Abstract
| Publisher Full Text
- 58.
Novais FO, Nguyen BT, Beiting DP, et al.:
Human classical monocytes control the intracellular stage of Leishmania braziliensis by reactive oxygen species.
J Infect Dis.
2014; 209(8): 1288–96. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 59.
Smeekens SP, van de Veerdonk FL, Joosten LA, et al.:
The classical CD14++ CD16- monocytes, but not the patrolling CD14+ CD16+ monocytes, promote Th17 responses to Candida albicans.
Eur J Immunol.
2011; 41(10): 2915–24. PubMed Abstract
| Publisher Full Text
- 60.
Passos S, Carvalho LP, Costa RS, et al.:
Intermediate monocytes contribute to pathologic immune response in Leishmania braziliensis infections.
J Infect Dis.
2015; 211(2): 274–82. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 61.
Campanelli R, Rosti V, Fois G, et al.:
CD14brightCD16low intermediate monocytes expressing Tie2 are increased in the peripheral blood of patients with primary myelofibrosis.
Exp Hematol.
2014; 42(4): 244–6. PubMed Abstract
| Publisher Full Text
- 62.
Turner JD, Bourke CD, Meurs L, et al.:
Circulating CD14brightCD16+ 'intermediate' monocytes exhibit enhanced parasite pattern recognition in human helminth infection.
PLoS Negl Trop Dis.
2014; 8(4): e2817. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 63.
Barbour JD, Jalbert EC, Chow DC, et al.:
Reduced CD14 expression on classical monocytes and vascular endothelial adhesion markers independently associate with carotid artery intima media thickness in chronically HIV-1 infected adults on virologically suppressive anti-retroviral therapy.
Atherosclerosis.
2014; 232(1): 52–8. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 64.
Zungsontiporn N, Tello RR, Zhang G, et al.:
Non-Classical Monocytes and Monocyte Chemoattractant Protein-1 (MCP-1) Correlate with Coronary Artery Calcium Progression in Chronically HIV-1 Infected Adults on Stable Antiretroviral Therapy.
PLoS One.
2016; 11(2): e0149143. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 65.
Ndhlovu LC, D'Antoni ML, Ananworanich J, et al.:
Loss of CCR2 expressing non-classical monocytes are associated with cognitive impairment in antiretroviral therapy-naïve HIV-infected Thais.
J Neuroimmunol.
2015; 288: 25–33. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 66.
Zhu Y, Davis S, Stephens R, et al.:
GEOmetadb: powerful alternative search engine for the Gene Expression Omnibus.
Bioinformatics.
2008; 24(23): 2798–800. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 67.
Banchereau R, Baldwin N, Cepika AM, et al.:
Transcriptional specialization of human dendritic cell subsets in response to microbial vaccines.
Nat Commun.
2014; 5: 5283. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 68.
Iwata M, Sandstrom RS, Delrow JJ, et al.:
Functionally and phenotypically distinct subpopulations of marrow stromal cells are fibroblast in origin and induce different fates in peripheral blood monocytes.
Stem Cells Dev.
2014; 23(7): 729–40. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 69.
Llaverias G, Pou J, Ros E, et al.:
Monocyte gene-expression profile in men with familial combined hyperlipidemia and its modification by atorvastatin treatment.
Pharmacogenomics.
2008; 9(8): 1035–54. PubMed Abstract
| Publisher Full Text
- 70.
Maouche S, Poirier O, Godefroy T, et al.:
Performance comparison of two microarray platforms to assess differential gene expression in human monocyte and macrophage cells.
BMC Genomics.
2008; 9: 302. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 71.
Hu X, Chung AY, Wu I, et al.:
Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways.
Immunity.
2008; 29(5): 691–703. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 72.
Wrzesinski SH, Fisher JL, Ernstoff MS:
Genetic profiles of plasmacytoid (BDCA-4 expressing) DC subtypes-clues to DC subtype function in vivo.
Exp Hematol Oncol.
2013; 2(1): 8. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 73.
Butchar JP, Cremer TJ, Clay CD, et al.:
Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion.
PLoS One.
2008; 3(8): e2924. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 74.
Boomgaarden I, Egert S, Rimbach G, et al.:
Quercetin supplementation and its effect on human monocyte gene expression profiles in vivo.
Br J Nutr.
2010; 104(3): 336–45. PubMed Abstract
| Publisher Full Text
- 75.
Karlsson KR, Cowley S, Martinez FO, et al.:
Homogeneous monocytes and macrophages from human embryonic stem cells following coculture-free differentiation in M-CSF and IL-3.
Exp Hematol.
2008; 36(9): 1167–75. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 76.
Szanto A, Balint BL, Nagy ZS, et al.:
STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells.
Immunity.
2010; 33(5): 699–712. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 77.
Poliska S, Csanky E, Szanto A, et al.:
Chronic obstructive pulmonary disease-specific gene expression signatures of alveolar macrophages as well as peripheral blood monocytes overlap and correlate with lung function.
Respiration.
2011; 81(6): 499–510. PubMed Abstract
| Publisher Full Text
- 78.
Strunnikova NV, Barb J, Sergeev YV, et al.:
Loss-of-function mutations in Rab escort protein 1 (REP-1) affect intracellular transport in fibroblasts and monocytes of choroideremia patients.
PLoS One.
2009; 4(12): e8402. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 79.
Ingersoll MA, Spanbroek R, Lottaz C, et al.:
Comparison of gene expression profiles between human and mouse monocyte subsets.
Blood.
2010; 115(3): e10–9. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 80.
Fuentes-Duculan J, Suárez-Fariñas M, Zaba LC, et al.:
A subpopulation of CD163-positive macrophages is classically activated in psoriasis.
J Invest Dermatol.
2010; 130(10): 2412–22. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 81.
Semnani RT, Keiser PB, Coulibaly YI, et al.:
Filaria-induced monocyte dysfunction and its reversal following treatment.
Infect Immun.
2006; 74(8): 4409–17. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 82.
Schmelzer C, Niklowitz P, Okun JG, et al.:
Ubiquinol-induced gene expression signatures are translated into altered parameters of erythropoiesis and reduced low density lipoprotein cholesterol levels in humans.
IUBMB Life.
2011; 63(1): 42–8. PubMed Abstract
| Publisher Full Text
- 83.
Smythies LE, Shen R, Bimczok D, et al.:
Inflammation anergy in human intestinal macrophages is due to Smad-induced IkappaBalpha expression and NF-kappaB inactivation.
J Biol Chem.
2010; 285(25): 19593–604. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 84.
Grigoryev YA, Kurian SM, Avnur Z, et al.:
Deconvoluting post-transplant immunity: cell subset-specific mapping reveals pathways for activation and expansion of memory T, monocytes and B cells.
PLoS One.
2010; 5(10): e13358. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 85.
Chen LY, Eberlein M, Alsaaty S, et al.:
Cooperative and redundant signaling of leukotriene B4 and leukotriene D4 in human monocytes.
Allergy.
2011; 66(10): 1304–11. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 86.
Liu F, Lei W, O'Rourke JP, et al.:
Oncogenic mutations cause dramatic, qualitative changes in the transcriptional activity of c-Myb.
Oncogene.
2006; 25(5): 795–805. PubMed Abstract
| Publisher Full Text
- 87.
Allantaz F, Cheng DT, Bergauer T, et al.:
Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression.
PLoS One.
2012; 7(1): e29979. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 88.
Santer DM, Wiedeman AE, Teal TH, et al.:
Plasmacytoid dendritic cells and C1q differentially regulate inflammatory gene induction by lupus immune complexes.
J Immunol.
2012; 188(2): 902–15. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 89.
Hulsmans M, Geeraert B, De Keyzer D, et al.:
Interleukin-1 receptor-associated kinase-3 is a key inhibitor of inflammation in obesity and metabolic syndrome.
PLoS One.
2012; 7(1): e30414. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 90.
Zhen A, Krutzik SR, Levin BR, et al.:
CD4 ligation on human blood monocytes triggers macrophage differentiation and enhances HIV infection.
J Virol.
2014; 88(17): 9934–46. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 91.
Irvine KM, Gallego P, An X, et al.:
Peripheral blood monocyte gene expression profile clinically stratifies patients with recent-onset type 1 diabetes.
Diabetes.
2012; 61(5): 1281–90. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 92.
Schenk M, Krutzik SR, Sieling PA, et al.:
NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy.
Nat Med.
2012; 18(4): 555–63. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 93.
Chauncey KM, Lopez MC, Sidhu G, et al.:
Bacillus anthracis' lethal toxin induces broad transcriptional responses in human peripheral monocytes.
BMC Immunol.
2012; 13: 33. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 94.
Henig N, Avidan N, Mandel I, et al.:
Interferon-beta induces distinct gene expression response patterns in human monocytes versus T cells.
PLoS One.
2013; 8(4): e62366. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 95.
Haniffa M, Shin A, Bigley V, et al.:
Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells.
Immunity.
2012; 37(1): 60–73. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 96.
Hou W, Gibbs JS, Lu X, et al.:
Viral infection triggers rapid differentiation of human blood monocytes into dendritic cells.
Blood.
2012; 119(13): 3128–31. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 97.
Ganesh K, Das A, Dickerson R, et al.:
Prostaglandin E2 induces oncostatin M expression in human chronic wound macrophages through Axl receptor tyrosine kinase pathway.
J Immunol.
2012; 189(5): 2563–73. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 98.
Chittezhath M, Dhillon MK, Lim JY, et al.:
Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression.
Immunity.
2014; 41(5): 815–29. PubMed Abstract
| Publisher Full Text
- 99.
Ismail N, Wang Y, Dakhlallah D, et al.:
Macrophage microvesicles induce macrophage differentiation and miR-223 transfer.
Blood.
2013; 121(6): 984–95. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 100.
Hyder LA, Gonzalez J, Harden JL, et al.:
TREM-1 as a potential therapeutic target in psoriasis.
J Invest Dermatol.
2013; 133(7): 1742–51. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 101.
Teles RM, Graeber TG, Krutzik SR, et al.:
Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses.
Science.
2013; 339(6126): 1448–53. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 102.
Wheelwright M, Kim EW, Inkeles MS, et al.:
All-trans retinoic acid-triggered antimicrobial activity against Mycobacterium tuberculosis is dependent on NPC2.
J Immunol.
2014; 192(5): 2280–90. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 103.
Shalova IN, Lim JY, Chittezhath M, et al.:
Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α.
Immunity.
2015; 42(3): 484–98. PubMed Abstract
| Publisher Full Text
- 104.
Martinez FO, Helming L, Milde R, et al.:
Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences.
Blood.
2013; 121(9): e57–69. PubMed Abstract
| Publisher Full Text
- 105.
Levine AJ, Horvath S, Miller EN, et al.:
Transcriptome analysis of HIV-infected peripheral blood monocytes: gene transcripts and networks associated with neurocognitive functioning.
J Neuroimmunol.
2013; 265(1–2): 96–105. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 106.
Radom-Aizik S, Zaldivar FP Jr, Haddad F, et al.:
Impact of brief exercise on circulating monocyte gene and microRNA expression: implications for atherosclerotic vascular disease.
Brain Behav Immun.
2014; 39: 121–9. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 107.
Kyogoku C, Smiljanovic B, Grün JR, et al.:
Cell-specific type I IFN signatures in autoimmunity and viral infection: what makes the difference?
PLoS One.
2013; 8(12): e83776. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 108.
Tilton JC, Johnson AJ, Luskin MR, et al.:
Diminished production of monocyte proinflammatory cytokines during human immunodeficiency virus viremia is mediated by type I interferons.
J Virol.
2006; 80(23): 11486–97. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 109.
Wu JQ, Sassé TR, Saksena MM, et al.:
Transcriptome analysis of primary monocytes from HIV-positive patients with differential responses to antiretroviral therapy.
Virol J.
2013; 10: 361. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 110.
Sharma S, Jin Z, Rosenzweig E, et al.:
Widely divergent transcriptional patterns between SLE patients of different ancestral backgrounds in sorted immune cell populations.
J Autoimmun.
2015; 60: 51–8. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 111.
Laudanski K, Miller-Graziano C, Xiao W, et al.:
Cell-specific expression and pathway analyses reveal alterations in trauma-related human T cell and monocyte pathways.
Proc Natl Acad Sci U S A.
2006; 103(42): 15564–9. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 112.
Reynolds LM, Taylor JR, Ding J, et al.:
Age-related variations in the methylome associated with gene expression in human monocytes and T cells.
Nat Commun.
2014; 5: 5366. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 113.
Mosig S, Rennert K, Büttner P, et al.:
Monocytes of patients with familial hypercholesterolemia show alterations in cholesterol metabolism.
BMC Med Genomics.
2008; 1: 60. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 114.
Däbritz J, Weinhage T, Varga G, et al.:
Reprogramming of monocytes by GM-CSF contributes to regulatory immune functions during intestinal inflammation.
J Immunol.
2015; 194(5): 2424–38. PubMed Abstract
| Publisher Full Text
- 115.
Elavazhagan S, Fatehchand K, Santhanam V, et al.:
Granzyme B expression is enhanced in human monocytes by TLR8 agonists and contributes to antibody-dependent cellular cytotoxicity.
J Immunol.
2015; 194(6): 2786–95. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 116.
Lee J, Breton G, Oliveira TY, et al.:
Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow.
J Exp Med.
2015; 212(3): 385–99. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 117.
Cho HJ, Shashkin P, Gleissner CA, et al.:
Induction of dendritic cell-like phenotype in macrophages during foam cell formation.
Physiol Genomics.
2007; 29(2): 149–60. PubMed Abstract
| Publisher Full Text
- 118.
Du X, Tang Y, Xu H, et al.:
Genomic profiles for human peripheral blood T cells, B cells, natural killer cells, monocytes, and polymorphonuclear cells: comparisons to ischemic stroke, migraine, and Tourette syndrome.
Genomics.
2006; 87(6): 693–703. PubMed Abstract
| Publisher Full Text
- 119.
Woszczek G, Chen LY, Nagineni S, et al.:
Leukotriene D4 induces gene expression in human monocytes through cysteinyl leukotriene type I receptor.
J Allergy Clin Immunol.
2008; 121(1): 215–21.e1. PubMed Abstract
| Publisher Full Text
- 120.
Liu PT, Stenger S, Li H, et al.:
Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response.
Science.
2006; 311(5768): 1770–3. PubMed Abstract
| Publisher Full Text
- 121.
Stegmaier K, Ross KN, Colavito SA, et al.:
Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation.
Nat Genet.
2004; 36(3): 257–63. PubMed Abstract
| Publisher Full Text
- 122.
Dower K, Ellis DK, Saraf K, et al.:
Innate immune responses to TREM-1 activation: overlap, divergence, and positive and negative cross-talk with bacterial lipopolysaccharide.
J Immunol.
2008; 180(5): 3520–34. PubMed Abstract
| Publisher Full Text



Comments on this article Comments (0)