Keywords
Transcriptomics, Bioinformatics, Placenta, Trophoblast, Diabetes, Pre-eclampsia, IUGR, trophoblast differentiation
This article is included in the Data: Use and Reuse collection.
Transcriptomics, Bioinformatics, Placenta, Trophoblast, Diabetes, Pre-eclampsia, IUGR, trophoblast differentiation
The second version of the manuscript includes additional validation results presented in an additional figure.
See the authors' detailed response to the review by Brian Cox
We aimed to make available via an interactive web-based application a collection of transcriptome datasets curated from the GEO public repository for their relevance to human placental development and pathology.
The placenta is a fetal organ indispensable for the establishment and maintenance of pregnancy. It connects the fetus to the maternal uterine wall via the umbilical cord, supplies nutrients and oxygen to the fetus, allows elimination of fetal waste, protects it from maternal infections and produces various hormones required for maintaining pregnancy1. The placental trophoblasts play critical roles for all of these activities by acting as an interface between fetus and mother2. In early embryogenesis, trophoblasts are the first cells differentiating from the fertilized egg, and eventually fusing with each other: a process transforming the monolayer cytotrophoblasts into syncytiotrophoblasts3. Several morphological and biochemical changes occur during this fusion process throughout pregnancy (known as trophoblast differentiation)4 to guarantee the development and appropriate functionality of the placenta1. Failure in placental formation, differentiation and function, particularly by trophoblast dysfunction, impacts fetal development and is associated with a wide range of pathologies, including gestational diabetes, hypertension, pre-eclampsia and intrauterine growth restriction (IUGR) of the fetus5. In addition, exposure of placenta to toxic compounds by a mother’s cigarette smoking alters the transcriptome of placental and fetal cells6. Indeed, maternal tobacco use causes various pregnancy-associated problems, including spontaneous fetal abortion, placental abruption, preterm birth, stillbirth, fetal growth restriction, and sudden infant death syndrome6,7.
With over 65,000 data deposited into the NCBI Gene Expression Omnibus (GEO), a public repository for functional genomics data, it can be challenging to identify datasets relevant to a particular research area. Indeed, GEO was primarily designed for the purpose of data archiving rather than data browsing and downstream analysis. Thus, we identified datasets from GEO particularly relevant to our interest in the development and pathology of the human placenta and uploaded them to the custom web-based Gene Expression Browser application (GXB) (http://placentalendocrinology.gxbsidra.org/dm3/landing.gsp), which provides seamless access to the data. GXB allows browsing and interactive visualization of large volumes of heterogeneous data. It also provides access to rich contextual information essential for data interpretation, such as: detailed gene information, relevant literature, study design, and sample information. In addition, the user can customize data plots by adding multiple layers of parameters, modify the sample order and generate links that can be shared via e-mail or used in future publications.
Thus we provide a resource enabling browsing of datasets relevant to placental development and pathology that offers a unique opportunity to identify genes that play a role in placental development/differentiation and are commonly modulated in pregnancy-associated diseases.
111 datasets, potentially relevant to the development and pathology of the human placenta, were identified in GEO using the following search query: “Homo sapiens[Organism] AND placenta[DESC]”. The majority of retrieved datasets were generated using Illumina or Affymetrix microarrays besides a few other commercial platforms.
The relevance of each one of the entries retreived with our query pertaining to “development and pathology of the human placenta” was assessed on a case-by-case basis. This process included reading the NCBI’s GEO-linked article and examining the list of samples profiled and their annotations as well as the study design. We selected for studies using primary placenta cells, placenta tissue or trophoblast cell lines comparing a) healthy with diseased samples or smoker vs. non-smoker or b) placenta cells in different differentiation/gestational stages. Finally we retrieved 24 curated datasets encompassing 759 transcriptome profiles, including 22 datasets generated using primary placenta cells and 2 datasets generated using trophoblast cell lines. Among the 24 datasets, there are 18 studies comparing control vs. diseased/smoker placenta cells, and 6 investigating transcriptomic changes during placental development and/or trophoblast differentiation. Among the many noteworthy datasets, several stood out, such as an extensive study comparing the diseased placentas with pre-eclampsia or unexplained IUGR vs. normal placentas (GSE24129)8. The datasets that comprise our collection are listed in Table 1.
For more information, see http://placentalendocrinology.gxbsidra.org/dm3/geneBrowser/list: PLAC1, placenta specific 1: CSH1, placental lactogen: XIST, X-inactive specific transcript NP: not published, no PubMed publication for this data set.
Once a final selection had been made, each dataset was downloaded from GEO in the SOFT file format. These files were then uploaded to the “placentaendocrinology” instance of GXB (http://placentalendocrinology.gxbsidra.org/dm3), an interactive web-based application developed at the Benaroya Research Institute, hosted on the Amazon Web Services cloud9. Information about samples and study design was also uploaded. Samples were then grouped based on relevant study variables and genes were ranked based on specified group comparisons. Details of the GXB software are described in a recent publication9. A tutorial for this software is also available online (https://gxb.benaroyaresearch.org/dm3/tutorials.gsp#gxbtut). GXB provides the user with a means to navigate and filter the dataset collection available at (http://placentalendocrinology.gxbsidra.org/dm3). Briefly, the datasets of interest can be quickly identified either by filtering with the pre-existing criteria listed in the column located in the left side of the dataset navigation window or by entering a query term in the search box located at the top of the window. Clicking one of the studies listed in the dataset navigation window opens a viewer, which is designed to support interactive browsing and graphic representations of the data in an interpretable format. This interface was developed to navigate ranked gene lists and to display expression results in a figure with a context-rich environment. Selecting a gene from the rank-ordered list located on the left side of the navigation window displays its expression values on the interactive plot. The drop-down menus located directly above the graphical display provide the user the following functions: a) Exporting the created graph as a portable network graphics (png) image or a csv file for performing a separate analysis. b) Changing ranking of genes in the list; this function allows the user to manipulate the ways of ranking the genes depending on his/her interest, or to include only the genes selected based on specific biological interest. c) Changing grouping of the samples (by using “Group Set” button); for example, the user can convert the groups created based on “cell type” to those on “disease type”. d) Identifying individual samples within a group using categorical or continuous variables (e.g., mode of delivery, ethnic group and age). e) Toggling between the bar chart view and a box plot view, with demonstration of the values as a single plot for each sample. Samples are split into different categories or groups whatever they are displayed in a bar chart or box plot. f) Providing a color legend for sample groups. g) Providing a color legend for the categorical information overlaid at the bottom of the graph. h) Selection of categorical information that is to be overlaid at the bottom of the graph. Using this function, the user can display, for example, gender or smoking status, at the bottom of the figure. The data without contextual information have no intrinsic utility. It is therefore important to capture and display the context information in the graph, so that the viewer can interpret the data shown in the graph. GXB provides functions for organizing the context information in the tabs located just above the graphical display. The tabs can be hidden to make more room for graphs, or be reappeared by clicking the blue “Show Info Panel” button located on the top right corner of the graphical display. Information on the genes selected from the list located in the left side of the graphical display is shown under the “Gene” tab. Information on the study for the demonstrated dataset is available under the “Study” tab. Detailed information of individual samples is provided under the “Sample” tab. Rolling the mouse cursor over a sample in either the bar chart or box plot view while displaying the “Sample” tab displays any clinical, demographic, or laboratory information available for that sample. Finally, the “Downloads” tab allows the advanced users to retrieve the original data for further analysis using other softwares/tools. It also provides all available sample annotation data along with the expression data. Other functionalities are provided under the “Tools” drop-down menu located in the top right corner of the user interface. Some of the notable functionalities available through this menu include: a) Annotations, which provides access to all the ancillary information about the study, samples and dataset organized across different tabs; b) Cross project view, which provides the ability to browse through all available studies while focusing on a the currently selected gene; c) Copy link, which generates a mini-URL encapsulating information about the display settings in use and that can be saved and shared with others (clicking on the envelope icon on the toolbar inserts the url in an email message via the local email client); d) Chart options, which gives user the option to customize chart labels.
We performed quality control checks on the datasets loaded on GXB by validating the expression of placental marker genes. Specifically, we used placenta specific 1 (PLAC1)10 and human placental lactogen (CSH1)11 (Table 1). As expected, all samples from the datasets we incorporated in our collection expressed these two placental markers (hyperlinked for easy access in Table 1), except some samples in GSE12216, GSE28551 and GSE10588, where CSH1 was not measured.
Gender specific expression of the XIST transcript (in the samples for which sex information was available) was also examined to determine its concordance with demographic information provided with the GEO datasets (Table 1). An overall accordance of a high XIST expression in female samples compared to a low XIST expression in male samples was determined in our entire dataset collection (Table 1). Based on our experience, concordance should be close to 100%. Levels of concordance closer to 50%, which were not observed here, would indicate a potential error with handling of samples during processing that may dramatically affect data analysis and interpretation (e.g. plate inversion).
We also verified and compared the differential expression using the Fold Change values obtained from GXB or the original manuscript. For this verification process, two datasets were selected GSE24129 and GSE30032. As shown in Figure 1, Fold Change values obtained from GXB correlate with those obtained from the original published article.
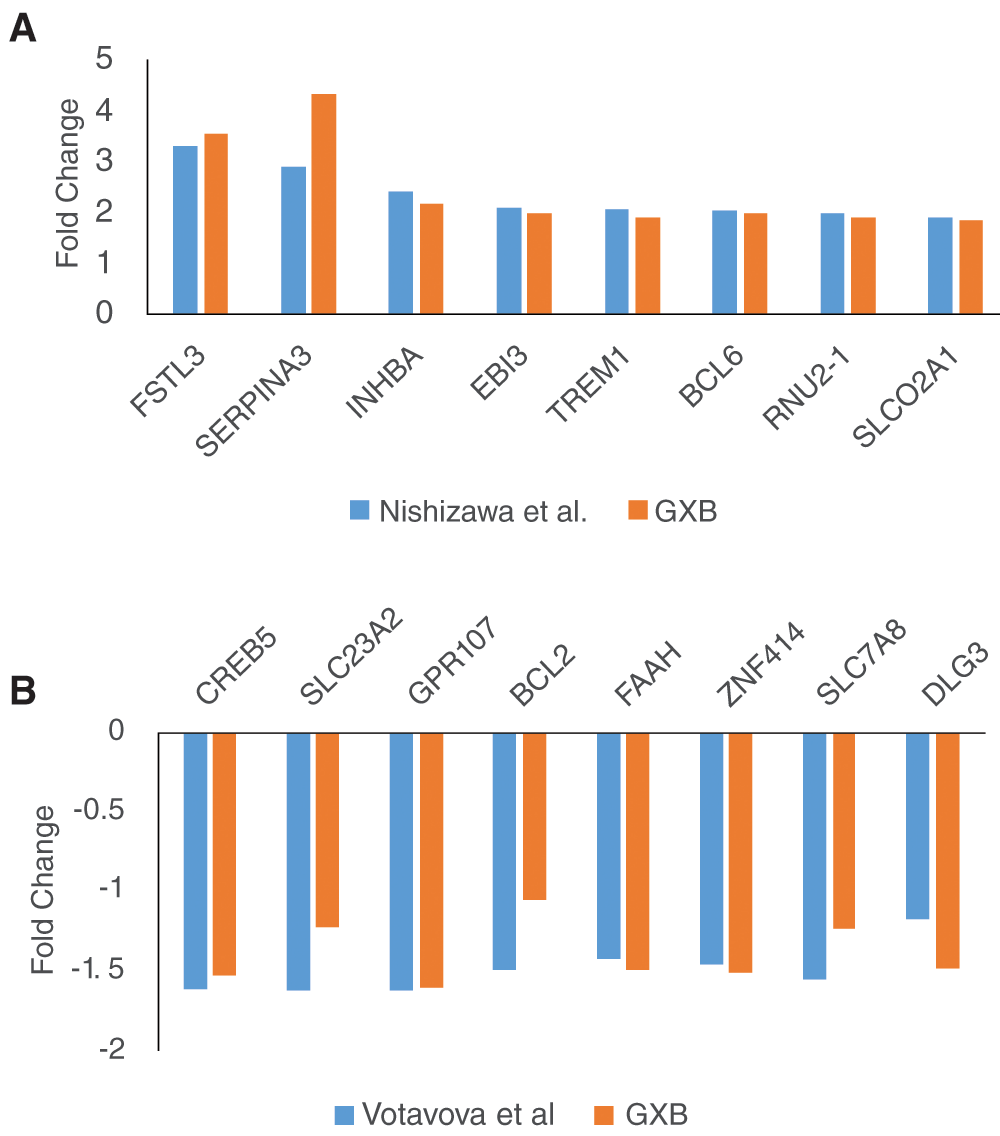
Two datasets were selected for validation of differential expression: GSE24129 (A) and GSE 30032 (B). The first 8 genes were selected from Table 2 in Nishizawa et al. (for GSE24129) and from supplemental material Table in Votavova et al. (for GSE30032).
All datasets included in our curated collection are available publically at the NCBI GEO website: http://www.ncbi.nlm.nih.gov/gds/. They were cited in our manuscript along with their GEO accession numbers (e.g. GSE24129). Signal files and sample description files for each uploaded GEO dataset can also be downloaded from the GXB tool under the “downloads” tab after accessing our dataset collection.
AM and TK conceived the theme for this dataset collection. AM, SB, SP and CQ contributed to the query, selection, loading and curation of datasets. DC and AM prepared the first draft of the manuscript. TK edited the manuscript. All authors were involved in the revision of the draft manuscript and have agreed to the final content.
All authors listed on this publication affiliated with the Sidra Medical and Research Center received support from the Qatar Foundation.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank all the investigators who decided to make their datasets publicly available by depositing them into GEO.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
References
1. Speake C, Presnell S, Domico K, Zeitner B, et al.: An interactive web application for the dissemination of human systems immunology data.J Transl Med. 2015; 13: 196 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 11 May 16 |
read | |
|
Version 1 09 Mar 16 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)