Keywords
Protein arrays, Whole-cell immunisation, Antibody profiling, Cross-reactivity,, Chicken IgY, Reference list, Secondary antibody, Detection antibody
This article is included in the Antibody Validations gateway.
Protein arrays, Whole-cell immunisation, Antibody profiling, Cross-reactivity,, Chicken IgY, Reference list, Secondary antibody, Detection antibody
Following the reviewers’ comments, this revised version of the manuscript now includes additional in silico sequence analysis assessing identified antibody cross-reactivities. The results and conclusion sections are now addressing the additional findings.
A new Table 5 and Supplementary Table S1 have been added showing results for all possible two-pair sequence alignment combinations presented as BLAST scores, SIM scores and E-values.
See the authors' detailed response to the review by Carsten Grötzinger
See the authors' detailed response to the review by Konrad Büssow
See the authors' detailed response to the review by Brigitte Hantusch
Secondary label-conjugated and non-conjugated detection antibodies are frequently used in a wide range of research applications. However, they are often affinity-isolated, polyclonal reagents that may lack the highest standard of antibody validation. The antibodies characterised in this study are a polyclonal anti-chicken IgY antibody produced in rabbit (31104, Thermo Fisher) and a polyclonal goat anti-rabbit IgG antibody conjugated with alkaline phosphatase (AP) (A3687, Sigma-Aldrich). Although the use of the rabbit anti-IgY antibody in the literature is limited, the goat anti-rabbit IgG AP has been extensively utilised for over 15 years1,2.
The research conducted in this laboratory examines complex antibody repertoires in humans and animals by means of protein arrays. Protein arrays are frequently used to profile antibody binding to human proteins in autoimmune disease3, cancer4 and in healthy individuals5. Other protein array applications include recombinant6 and hybridoma-derived7 antibody characterisation studies. This article investigates the cross-reactivity of a rabbit anti-chicken IgY and an alkaline phosphatase-conjugated goat anti-rabbit IgG, which were used for the profiling of IgY antibody responses to human antigens in chickens immunised with human cancer cells. The protein array technology applied here, developed by Büssow and colleagues8, is comprised in its current version of a fully annotated set of 7,390 distinct human proteins, that may serve as potential antigens. The aim of this study is to define a cross-reactivity reference list for the two described secondary antibodies, which can then be used to eliminate non-specific binders from ongoing chicken IgY profiling studies. Furthermore, publication of the cross-reactivity reference list provides a valuable resource of potential false-positive binders to researchers using the same antibodies. may support other researchers using these antibodies in the evaluation of their experiments.
Rabbit anti-chicken IgY (H+L) secondary antibody (Thermo Fisher Scientific, Product number 31104, Lot code PK19380211) is a polyclonal antibody that targets the variable heavy and light chains of chicken IgY immunoglobulins (Table 1). The antibody was isolated from the serum of the antigen-immunised rabbit through immunoaffinity chromatography using antigen coupled to agarose beads. The antibody was added to the protein array at a 1/1,000 dilution in 2% (w/v) bovine serum albumin (BSA, Sigma-Aldrich, A2153) in tris-buffered saline (TBS, Trizma® Base, Sigma-Aldrich, T6066 and sodium chloride, Fisher Scientific, S/3160/68) with 0.1%, v/v, Tween 20 (Sigma-Aldrich, P1379).
Alkaline phosphatase-conjugated goat anti-rabbit IgG (whole molecule) (Sigma-Aldrich, Product number A3687, Lot code SLBJ6146V) is a polyclonal antibody that targets all rabbit IgGs (Table 1). The antibody was isolated through immunospecific purification of antisera from a rabbit IgG-immunised goat. Following isolation, the anti-rabbit IgG was conjugated to alkaline phosphatase using glutaraldehyde-based cross-linkage. The antibody was added to the protein array at a 1/1,000 dilution in 2% (w/v) BSA in tris-buffered saline (TBS) with 0.1%, v/v, Tween 20.
Unipex protein arrays were obtained from Source Bioscience Life Sciences (Nottingham, UK). The Unipex arrays comprise of 15,300 fully annotated E. coli clones expressing a total of 7,390 distinct in-frame ORF human recombinant proteins. The Unipex proteins are immobilized under denaturing conditions directly on the PVDF membrane surfaces exposing linear sequence epitopes ideally suited for epitope mapping, antibody profiling and antibody cross-reactivity analyses. The details of protein arrays utilised in this study are provided in Table 2. For general information on Unipex protein arrays please refer to: (http://www.lifesciences.sourcebioscience.com/media/290406/sbs_ig_manual_proteinarray_v1.pdf).
Antibody cross-reactivity was assessed using Unipex protein arrays. The detailed experimental protocol is provided in Table 3. Briefly, secondary rabbit anti-chicken IgY and goat anti-rabbit IgG AP were validated in preparation for a chicken IgY antibody profiling experiment of a chicken immunised with human cancer cells. Protein arrays were probed with secondary antibodies in the absence of IgY-containing chicken serum, as described in Table 3. Signal generation for array-bound secondary antibodies was obtained using AttoPhos AP fluorescent substrate system (Promega, S1001) diluted 1 in 8 in AP buffer (1mM MgCl2, Sigma-Aldrich, M4880 and 100mM Tris base, pH 9.5). Protein array image acquisition was conducted using a Fuji scanner Fla5100. Positive signals were localized according to the manufacturer’s protocol. Briefly, array proteins were spotted in duplicate in a 3×3 square pattern. The centre spot of each square being a guide dot surrounded by eight flanking protein spots. Each protein was spotted around the navigation dot in one of four predetermined patterns (see Figure 1b). Varying background intensities were controlled by adjusting brightness and contrast of the image using Visual Grid software (GPC Biotech) to allow best possible scoring conditions. The degree of signal intensity was evaluated for each protein pair with the value 1 corresponding to a weak signal, value 2 corresponding to a moderate signal and value 3 corresponding to a strong signal. The x- y- coordinates of each positive pattern were merged with the Unipex protein database provided by the manufacturer (Source Bioscience) resulting in identification of GenBank and UniGene ID’s for each positive signal. This is a commonly used method for scoring signal intensities as previously shown by this group4 and others9,10. Protein annotations were retrieved from the Unipex database provided by the manufacturer and updated using the National Cancer Institute’s UniGene CGAP Gene Finder tool (http://cgap.nci.nih.gov/Genes/GeneFinder).
To investigate whether antibody binding to protein arrays was due to epitope similarities between the animal immunogens used to produce the secondary antibodies and the human proteins on the arrays we performed a comparative analysis as follows. Sequences of human antigens on the array bound by the secondary antibodies were obtained from the PubMed website (http://www.ncbi.nlm.nih.gov/protein/) using IDs present in the Unipex protein database and compared to chicken immunoglobulin proteins [Ig lambda chain C region (NCBI Accession: P20763.1), Ig lambda chain V-1 region (NCBI Accession: P04210.1), immunoglobulin Y heavy chain constant region (NCBI Accession: XP_015130394.1) and immunoglobulin Y heavy chain variable region (NCBI Accession: ADF29959.1)], as well as to rabbit immunoglobulin proteins [Ig gamma chain C region (UniProtKB: P01870), immunoglobulin heavy chain VDJ region, partial (NCBI Accession: AAA51320.1), Ig lambda chain C region (UniProtKB: P01847.2) and Ig lambda chain variable region, partial (NCBI Accession: AAA31364.1)]. For antigen similarity comparisons, sequence similarities were analysed using BLAST. Non-intersecting protein sequence alignments were analysed using the local similarity program SIM adjusted to the BLOSUM62 comparison matrix to ensure amino acid complementarity of linear B-cell epitopes as previously shown11. The threshold for sequence similarity was set to BLAST E-values below 1 × 10-10 and SIM score values above 50.
Probing protein arrays with antibodies allows the assessment of their specificity and cross-reactivity across a large numbers of potential antigens in parallel12,13. Here we investigated the cross-reactivity of a secondary rabbit anti-chicken IgY and a goat anti-rabbit IgG labelled with AP, using a single set of human protein arrays in the absence of chicken serum. We identified a total of 63 binding events, of which 61 corresponded to unique proteins (Table 4). The identified positive signals varied in strength, as shown in Figure 1, with intensity 3 being the strongest and 1 the weakest. Five of the identified signals were scored as intensity 3, twelve signals were scored as intensity 2 and remainder scored as intensity 1. The original protein array images are shown in Figure S1 and Figure S2 (Supplementary material) and protein array images with highlighted positive signals, which correspond the cross-reactive proteins listed in Table 4, are shown in Figure S3 and Figure S4 (Supplementary material).
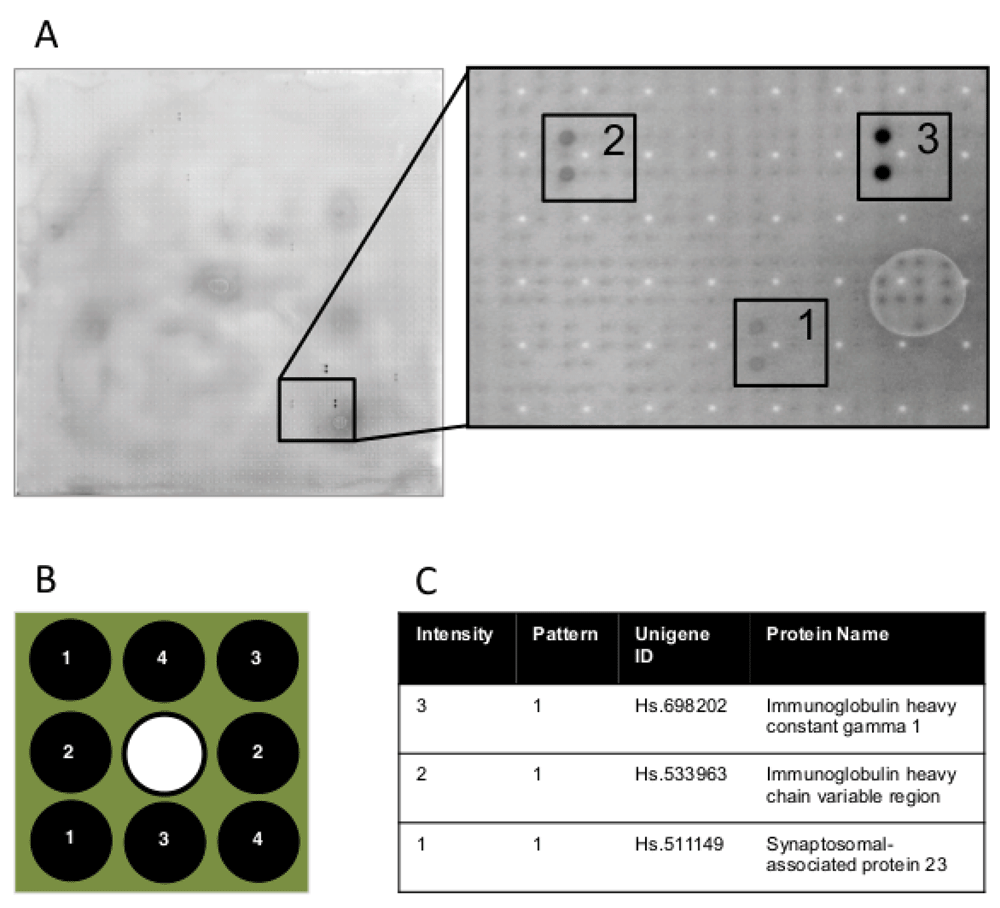
(A) Image of a whole protein array and a representative section illustrating antibody-antigen binding at three different signal intensities; 3 = strong, 2 = intermediate and 1 = weak. (B) The proteins are arranged in a 3×3 pattern on the array and all proteins are arrayed twice and appear as duplicate spots in a particular pattern within a block after a successful hybridization. (C) Description of proteins chosen as examples provided on the representative array image above; signal intensities, patterns, Unigene IDs and protein names are listed.
The 61 identified proteins comprised of a wide range of human proteins, including immunoglobulins, as well as a variety of nuclear, cytoplasmic and cell-membrane proteins with a diverse range of functions (Table 4). In order to identify shared epitopes that could explain the observed antibody cross-reactivity, and to deduce the origin of the non-specific binding to either of the two tested antibodies, we investigated sequence similarities between the human proteins and the immunogens used to produce the antibodies. We conducted a linear (BLAST) and a segmented (SIM) in silico sequence analysis of chicken IgY and rabbit IgG immunoglobulins against 61 array-identified human proteins as detailed in supplementary Table 1. In total, 5 proteins met the BLAST threshold criteria of E-values below 1 × 10-10, as well as the SIM threshold criteria of scores above 50. A further 9 proteins met the SIM threshold, but they did not meet the BLAST criteria (Table 5).
All 5 proteins that met both threshold criteria belong to the immunoglobulin class of proteins, four being variable regions of Ig heavy chains and one a heavy constant gamma chain. The in silico sequence analysis revealed the highest sequence similarity to Ig heavy variable and constant chains, respectively, of both, the chicken IgY and rabbit IgG in all cases (Supplementary Table 1) making it impossible with this approach to deduce the origin of the cross-reactivity.
14 most similar human proteins detected in this study, prioritised by sequence similarity.
| Human protein | Chicken Immunoglobulins | Rabbit Immunoglobulins | Signal Intensity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BLAST overlaps* | Highest BLAST E-value | SIM overlaps* | Highest Score | BLAST overlaps* | BLAST | SIM overlaps* | Highest Score | ||
| Clone SFV019_2F0 5H immunoglobulin heavy chain variable region | 1 | 1.00E-48 | 2 | 393 | 2 | 3.00E-47 | 4 | 356 | 3 |
| Clone IgA-MZ-aa42c-2 immunoglobulin alpha heavy chain variable region (IgA) | 1 | 4.00E-30 | 2 | 210 | 1 | 1.00E-29 | 1 | 194 | 3 |
| IGH mRNA for immunoglobulin heavy chain VHDJ region, partial cds, clone:TRH1-16 | 2 | 2.00E-34 | 1 | 218 | 1 | 1.00E-41 | 2 | 291 | 3 |
| Immunoglobulin heavy constant gamma 1 (G1m marker) | 1 | 3.00E-13 | 1 | 86 | 2 | 2.00E-125 | 2 | 890 | 3 |
| Clone IP80 immunoglobulin heavy chain variable region | 1 | 3.00E-44 | 1 | 313 | 1 | 3.00E-49 | 2 | 357 | 2 |
| Enhancer of polycomb homolog 1 (Drosophila)e | 0 | BT | 1 | 51 | 0 | BT | 0 | BT | 2 |
| Inverted formin, FH2 and WH2 domain containing | 0 | BT | 0 | NA | 0 | BT | 1 | 51 | 1 |
| Chromosome 10 open reading frame 114 | 0 | BT | 1 | 51 | 0 | BT | 0 | BT | 1 |
| RUN and SH3 domain containing 2 | 0 | BT | 1 | 52 | 0 | BT | 0 | BT | 1 |
| Single stranded DNA binding protein 4 | 0 | BT | 1 | 56 | 0 | BT | 0 | BT | 1 |
| Rho GTPase activating protein 33 | 0 | BT | 1 | 54 | 0 | BT | 0 | BT | 1 |
| Splicing factor 3b, subunit 4, 49kDa | 0 | BT | 1 | 61 | 0 | BT | 0 | BT | 1 |
| Protein phosphatase 1, regulatory subunit 26 | 0 | BT | 1 | 59 | 0 | BT | 0 | BT | 1 |
| Alpha tubulin acetyltransferase 1 | 0 | BT | 1 | 50 | 0 | BT | 0 | BT | 1 |
‘BT’ signifies ‘Below Threshold Value’. Threshold values were BLAST E-values below 1 × 10-10 and SIM values above 50. ‘NA’ signifies ‘No significant similarity was detected’
*BLAST and SIM overlaps indicate the number of sequence categories meeting threshold criteria for similarity as shown in Supplementary Table 1.
The 9 proteins that met only the SIM threshold criteria belong to a wide range of protein classes, however, none of those proteins belongs to the immunoglobulin class of proteins. In silico sequence analysis revealed that 8 of those proteins have a high local sequence similarity to the chicken immunoglobulin Y heavy chain constant region, but not to any other chicken and rabbit Ig regions. The analysis revealed further, that Inverted Formin, FH2 and WH2 Domain Containing (INF2) showed high local similarity exclusively to the rabbit Ig gamma chain constant region (Supplementary Table 1).
This work illustrates the cross-reactivity of an antibody-based detection system for IgY binding. The polyclonal anti-IgY rabbit antibody in combination with an anti-rabbit IgG alkaline phosphatase-conjugated antibody was shown to bind to 61 human proteins present on Unipex protein arrays comprising of 7,390 human proteins. Characterisation of this cross-reactivity provides a ‘false-positive’ database for future chicken antisera characterisation on protein array systems not limited to the Unipex protein array used here. These results, in combination with ‘false-positives’ from earlier research investigating antibody cross-reactivity by this group12 and others13 may provide valuable information for future protein array-based experiments. Reference lists provided by such experiments would be further strengthened by arrays that include additional portions of the human proteome and/or post-translational modifications. Using antibodies that have been extensively characterised on protein arrays will reduce the risk of identifying irrelevant cross-reactive secondary antibody binding to the array as a host-antigen response.
It is important to note that the current study was a one-off experiment and repeat experiments may increase the reliability of the data. The reproducibility of the binding events identified in this study was further warranted by evaluating each protein in two discrete positions on the array. Of the 63 binding events, five were scored as intensity 3, twelve were scored as intensity 2 and the remainder were intensity 1. While the assay is unable to conclusively distinguish the precise cause of the differences in signal intensities, it can be assumed to be due to variations in antibody affinity and avidity, the availability of the epitope for binding, and protein concentrations on the array. A follow-up quantitative Western blot analyses and titration experiments would help further to shed more light into differences in antigen-binding kinetics.
The secondary antibodies utilized in this study are polyclonal, isolated by immunoaffinity chromatography. The presented cross-reactivity reference list may, therefore, show some variation when a different lot of the antibody is used. We have previously shown that conditions applied during affinity chromatography may affect specificity14. When assessing protein array images, we found a considerable discrepancy in background intensity of array part 1 and 2. It is important to highlight that the part 1 and part 2 of the array are generated from distinct clone libraries of different tissue origin. Part 1 of the array was generated from human brain tissue using a pQE30NST vector, whereas part 2 of the array was generated from different sources of tissue, including T cells and lung tissue, using a pQE80LSN vector. The tissue origin and the utilised bacterial vector are potential contributing factors for the variances in background noise.
Since both antibodies were used as a pair in this study, it was not possible to directly deduce the exact cross-reactivity profile for each individual antibody. We have therefore taken an in silico sequence analysis approach and we found that five of the identified proteins were of the immunoglobulin class of proteins with very high sequence similarities to both, the chicken IgY and the rabbit IgG immunoglobulins. Such cross-reactivity is not surprising considering that the antibodies are polyclonal and the immunogens were immunoglobulins of both hosts. In addition, the data sheet provided with the anti-chicken IgY antibody produced in rabbit (31104, Thermo Fisher) has specified that this antibody may cross-react with immunoglobulins from other species. The data sheet for the goat anti-rabbit IgG AP antibody (A3687, Sigma-Aldrich) has specified binding to all rabbit immunoglobulins. The in silico sequence analysis revealed furthermore 8 proteins with high sequence similarity to chicken IgY heavy chain constant region and one protein with high sequence similarity to rabbit Ig gamma chain constant region. In order to tackle this issue experimentally, a single labelled antibody should be tested on its own in future experiments. Furthermore, if a non-labelled antibody is to be tested, two experiments should be performed, one with a labelled and non-labelled antibody pair such as demonstrated in this study, and one additional experiment with the labelled antibody alone, thereby allowing allocation of exact cross-reactivates by simply subtracting ‘false-positives’ from both sets of results.
In conclusion, the antibodies tested in this study showed cross-reactivity to unrelated human proteins as well as to human immunoglobulin proteins, which are homologous to the original immunogens. Despite the identified non-specific binding, the tested antibodies are suitable for use in protein array experiments as the cross-reactive binding partners can be readily excluded from further analysis. As both antibodies were used as a pair in this study, the possibility to deduce the exact cross-reactivity profile for each individual antibody may be limited. However, the cross-reactivity reference list provided in this paper can be further utilised to validate research using those antibodies in applications other than protein arrays.
ROK and GSK designed the study, DL performed the protein array experiments and GSK conducted data analysis. GSK conceived and DL performed in silico sequence analyses. GSK wrote and DL and ROK critically reviewed and edited the article. All authors have agreed to the final content of the manuscript.
This material is based upon works supported by the Irish Cancer Society Research Fellowship Award CRF10KIJ (GSK), the Science Foundation Ireland under CSET Grant no. 10/CE/B1821 and the Enterprise Ireland Dairy Processing Technology Centre award.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Table S1. Chicken IgY, rabbit IgG and array-identified human proteins in silico sequence analysis data. In order to investigate whether potential shared epitopes exist between immunogens (chicken IgY, rabbit IgG) used to produce the secondary antibodies tested in this study and the human proteins bound non-specifically by those secondary antibodies on protein arrays, we analysed sequence similarities using BLASTP and the local similarity program SIM. We used sequences of protein IDs provided by the Unipex protein database and compared those to chicken and rabbit immunoglobulin proteins (Methods section, Epitope analysis). The Table S1 shows the results for all possible two-pair sequence alignment combinations presented as BLAST scores, E-values and SIM scores.
Figure S1. Unipex 1 pt.1 protein array image. Original image of protein array (Number 633.4.730) probed with rabbit anti-chicken IgY and alkaline phosphatase-conjugated goat anti-rabbit IgG, visualised using AttoPhos AP Fluorescent Substrate.
Figure S2. Unipex 2 pt.1 protein array image. Original image of protein array (Number 634.5.737) probed with rabbit anti-chicken IgY and alkaline phosphatase-conjugated goat anti-rabbit IgG, visualised using AttoPhos AP Fluorescent Substrate.
Figure S3. Unipex 1 pt.1 protein array image with highlighted positive signals. Cross-reactive proteins listed in Table 4 are highlighted corresponding to their intensity as red (intensity 3 = strong), green (intensity 2 = intermediate) and yellow (intensity 1 = weak) circles.
Figure S4. Unipex 2 pt.1 protein array image with highlighted positive signals. Cross-reactive proteins listed in Table 4 are highlighted corresponding to their intensity as red (intensity 3 = strong), green (intensity 2 = intermediate) and yellow (intensity 1 = weak) circles.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 23 May 17 |
read | ||
|
Version 1 18 Jan 16 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)