Keywords
breast cancer, BRCA1 gene, SNP, novel mutation, missense variants, heterozygous, Sudanese patients
breast cancer, BRCA1 gene, SNP, novel mutation, missense variants, heterozygous, Sudanese patients
I would like to thank the Referees for their agreement to participate in sharing knowledge and experience by providing valuable notes and recommendations that to be considered for better scientific improvements to our version 1. In response to these recommendations, a reference citation has been added for the African Breast cancer statistics. In addition, Figure 1 has been replaced with a figure with better resolution.
See the authors' detailed response to the review by Khalid Dafaallah Awadelkarim
Breast cancer (BC) is a very serious issue worldwide, and is one of the leading causes of death in women today. In the US, it was estimated that there were approximately 232,670 new cases of BC and 40,000 BC deaths in 2014, and the number increased with 13,990 new cases and 450 deaths in 20161,2. In Africa, in 2012, the rate was about 94,000 women with BC, which resulted in 48,000 deaths3, and studies in Africa have described a poor outcome with a late diagnosis, due to the aggressiveness of the disease and the absence of screening programs4–7.
In Sudan, BC occurs at the highest frequency among women compared to other types of cancer8–13. In a hospital-based statistical report in Sudan13, BC was found to be the most commonly diagnosed malignant tumor and was characterized by early onset and bad prognosis. The report showed invasive ductal carcinoma to be the predominant type (82%), and 74% of patients were <50 years old with an advanced disease stage, indicating that most cases remain undiagnosed for long periods8,9,13–15. BRCA1 (OMIM_113705) was mapped in 1994 and subsequently cloned. It is located on chromosome 17 region 2, band 1 (17q21), which is responsible for encoding 1863 amino acids16. Since 1995, the BRCA1 tumor suppressor protein has been found to arrest cell proliferation, play an important role in the repairing process of DNA damage, and was suggested to have a role in cell cycle regulation through interacting directly or indirectly with other regulatory molecules17–22. BRCA1 when it is altered becomes deficient, and such loss of function mutates the protein, which not only perturbs chromosomal integrity and genome stability, but increases the mutation rate of other genes23–26. Therefore, it has been proposed that BRCA1 doesn’t directly initiate cancer formation, but enhances the process by making the affected cells highly susceptible to malignant transformation27,28. Germline mutations in BRCA1 are responsible for a large proportion of inherited predispositions for BC, and individuals that carry an inherited mutation in the BRCA1 gene have a significant risk (46–87%) for developing BC by 70 years of age29–34. Regionally among Africa and locally in Sudan, BRCA1-associated BC has been identified among premenopausal women6,9,35. This indicates the highly susceptible nature of such a mutation to enhance cancer development earlier during fertile reproductive women. African scientific literature has greatly studied the risk of BC resulting from reproductive factors (such as early menarche, late menopause, and sex hormones), but has not explored much into the genetic predisposition of the disease6,36–38.
About 13108 SNPs have been reported within human BRCA1, of them 1608 were reported as missense variants and about 151 have been identified to be pathogenic, according to the SNP database at NCBI (http://www.ncbi.nlm.nih.gov/snp). Most of these mutations resulted from small insertions/deletions, leading to frameshift, stop codon or nonsynonymous missense substitution, deletion, duplication and disruption of splice sites, resulting in a nonfunctional protein39. The SNP variant rs1799950 was observed to have a negative association with BC, by favoring more frequently the control groups than BC patients40,41. This SNP was tested among familial BRCA1 carriers for BC risk association, and a significant association was found within affected families42. When the SNP was haplotype-homozygous within affected families, the risk was increased, as in the case of sporadic risk association study43. The hetero-homozygosity nature of this mutation has been noticed within different studies, and some studies found the heterozygous variant more frequently within controls, hence it was adversely associated with BC, while in another study, the homozygous variant was found more frequently within BC patients40,43. In addition, heterozygosity was functionally assessed among monoallelic BRCA1 mutation carriers of rs1799950, and the results showed that such an alteration could permit variation in protein expression and activity in a haploinsufficient way, which could alter the cell’s normal behavior and result in tumor transformation by enhancing tumorigenesis. Such variation conferred by one mutated copy suggests that the wild-type copy alone is not capable of compensating the loss of the other wild allele44,45. Turkovic et al. found that such a haplotype association was noticed more frequently within deleterious mutation carriers; however, this was observed in a small sample size46. In addition, some studies have found rs1799950 to be associated with early-onset prostate cancer47–49.
Two genetic studies have been conducted in Sudan concerning BRCA1. One was a survey of 2370 students at a girl’s secondary school in Northern Sudan-Marawi, in which the study divided 67 students into two groups (47 students with a family history of BC and 20 with unaffected families) to analyze BRCA1 and BRCA2 mutations. In the first group, which was 2.37% of responders, the frequency of mutations was higher for BRCA1, and most mutations were within exon 11. The study continued to recommend further assessments of this region in subsequent local projects, and this formed the basis of our primer (1 and 2) selection within the present study10. The other study, from central Sudan, investigated 34 early onset premenopausal women patients diagnosed with BC (<40 years) and one male patient. The study identified 60 mutations in these patients, five of which were deleterious, affecting the outcome protein9.
From the same region within central Sudan, early onset BC premenopausal women have also been investigated for BRCA1 point mutations. The findings revealed the presence of one deleterious variant, 24 neutral variants and eight variants of unknown significance, within which two novel variants were discovered35.
Since there have been scarce genetic studies conducted highlighting genetic characteristic and familial risk status of BC patients in Africa (38) our aim was to screen for the type and spectrum of germline mutations in BRCA1 by focusing on three regions within the gene, two within exon 11 and one within exon 20, using sequencing, and to further assess the detected variants using in silico analysis tools. These regions and their selections were based on the quality of available primers (e.g. best GC content, adequate length, according to previous literature50), previous local research findings revealing frequent mutations within exon 119,10, and the cost.
This study was carried out in March 2015 at the Radiation and Isotope Center in Khartoum. 2–3 mls of blood were collected randomly from 45 patients diagnosed with BC who attended the center for treatment and follow-up (no other inclusion/exclusion criteria were relevant), using sterile EDTA-K3 vacutainer and kept at -20°C. All the patients were aged between 27 and 80 years old, with a mean of 45.9 years. Early onset cases were more frequent than late onset: 25 (55.6%) cases with early onset, with a mean of 36.6 years; and 20 (44.4%) cases of late onset with a mean of 57.4 years. Multiparity was high in 30/45 (66.6%). Six cases reported a family history of BC (13.3%), abortion was detected in 10 cases (22.2%). Of the presented histotypes that were available to us (16 cases unknown), ductal type tumor was more frequent and shown in 22 cases (48.8%), then lobular in five cases and mucinous in two cases. In addition, five cases showed distal cancer metastasis; lung and bone were frequent secondary sites. Right side tumor originated in 20 cases (44.4%), while the left side tumor was found in 15 cases (33.3%), and four cases presented with bilateral BC (6 cases unknown) (see Table 1).
*Comprising both Khartoum 16 cases and AlGezirah 5 cases.
| Variable | Frequency, n (%) (n=45) | |
|---|---|---|
| Onset | Early (≤45 years) Late (≥46 years) | 25 (55.6) 20 (44.4) |
| Family history | Breast cancer Other cancer No family history of any cancer | 6 (13.3) 5 (11.1) 34 (75.6) |
| Parturition | Multiparous Nulliparous Primiparous | 30 (66.7) 13 (28.9) 2(4.4) |
| History of abortion | Yes No | 10 (22.2) 35 (77.8) |
| Marital status | Currently married Single Previously married | 41 (91.1) 3 (6.7) 1 (2.2) |
| Tribe | Ja'alya Shaygeya Dnagla Noba Rezaigat Other | 5 (11.1) 5 (11.1) 4 (8.9) 3 (6.7) 3 (6.7) 25 (55.5) |
| Geographical region | Central Sudan* Western Sudan Northern Sudan Eastern Sudan | 21 (46.7) 15 (33.3) 6 (13.3) 3 (6.7) |
| Tumor site | Unilateral Bilateral Unknown | 35 (77.8) 4 (8.9) 6 (13.3) |
This study was conducted under the guidelines and approval of the Research Ethics Committee of Sudan Ministry of Health – Khartoum state. All participants provided oral informed consent to participate in the study. Oral informed consent was obtained as opposed to written consent, due to the literacy levels of the patients and the time limited interaction between researchers and patients at the hospital.
DNA was extracted using the salting-out method51 for 45 patients samples. In addition, proteinase K was used to enhance WBC membrane breakdown at 56°C for 1 hour. For PCR, three previously published pairs of primers50 were used to amplify three regions within the BRCA1 gene. All the three primers were selected for their quality performance, optimal size and GC content, after being assessed with Oligoanalyzer tool 3.1 (https://www.idtdna.com/calc/analyzer). These primers were synthesized by Macrogen Incorporation (Seoul, South Korea; Table 2). Annealing temperatures were adjusted on several runs (Table 2). Maxime PCR PreMix Kit i-Taq 20 μl (INTRON Biotechnology, South Korea) was used for PCR - 15 ul distilled water, 3 ul sample DNA (30 ng/ul; as checked by NanoDrop 1000) and 1 ul of the final concentration of each primer (10 pmol/µl forward and reverse). PCR mixture was subjected to initial denaturation step at 96°C for 5 minutes; followed by 35 cycles of denaturation at 96°C for 30 seconds, primer annealing at 50 or 55°C depending on the set used, for 30 seconds; followed by a step of elongation at 72°C for 60 seconds; the final elongation was at 72°C for 10 minutes50. After PCR amplification, the PCR products (442, 271 and 401bp) were checked by 2% gel electrophoresis at 100 V for 30–45 min (Figure 1).
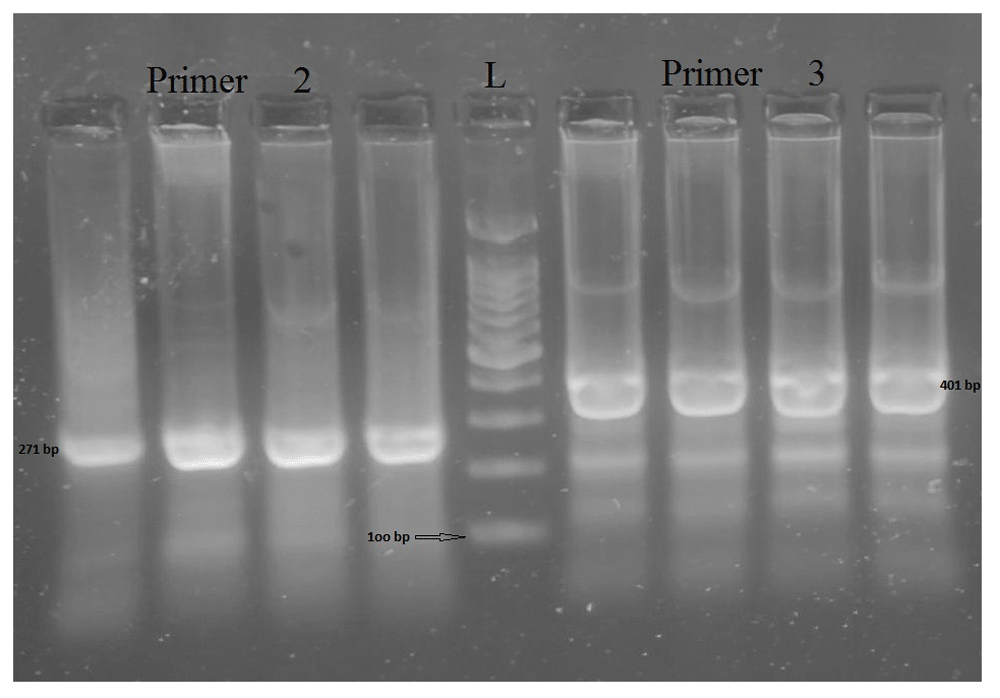
Left-side, PCR amplicons of primer 2, size 271 bp; right-side, PCR amplicons of primer 3, size 401 bp; L, ladder of 100bp each.
F: forward; R: reverse; bp: base pair; CDS: coding sequence; V1: variant one.
The product size of the first and last primers were checked and assessed using Serial Cloner version 2.6.1 (http://serialbasics.free.fr/Serial_Cloner.html) on the known nucleotide database accession gene for BRCA1 (NG_005905) with the whole sequence size of 81189bp, both forward and reverse of each one have been found to determine regions that cover coding and non-coding sequences.
The NCBI RefSeqGene NG_005905, which represents the whole BRCA1 gene, and the transcript variant 1 NM_007294 mRNA, which comprises mainly the coding sequences of the gene. The first sequence was used to check that all three primers amplicons within the BRCA1 gene region, while the second sequence was used for assessing all the three primers amplicons within the BRCA1 coding sequence. The gene sequence of BRCA1 has marked all the primers set amplicons to be within the gene region sequence. The transcript variant 1 (NM_007294) mRNA has marked (only 367 and 86bp) nucleotide sequences within primers sets 1 and 3 amplicons, respectively, to be within coding sequences, and the whole set of primer 2 amplicon (271bp) was within the coding sequence of the BRCA1 gene (Table 2).
The PCR products of the 10 best bands yielded from the patient samples for each primer set, a total of 30 resulted amplicons, were sent for Sanger dideoxy sequencing. Partial standard sequencing for the three regions within the gene, including both forward and reverse nucleotide sequencing, was performed by Macrogen Company (Seoul, South Korea), using the same pairs of primers.
Sequence analysis. The sequence results for the 30 sequence chromatogram files were viewed by FinchTV program version 1.4.052, which was used to check both nucleotide sequences of the patients forward and reverse sequences to be free of errors. Any errors were excluded during processing. The Basic Local Alignment Search Tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to assess nucleotide and protein sequence similarities53. In ExPASy translate tool - SIB Bioinformatics Resource Portal, the gene sequences were translated into amino acid sequences54. For primers 2 and 3, the BRCA1 nucleotide sequences from the patients, with their translated proteins, underwent multiple sequence alignment using BioEdit software version 7.0.9.055. Multiple sequence alignment included the reference sequence with the highest similarity, as obtained by BLAST (RefSeq transcript mRNA - NM_007294 transcript variant 1), two additional nucleotide sequences (NM_007297.3, transcript variant 3 mRNA and JN686490.1; Figure 2A), and the gene sequence NG_005905, which is mainly the sequence between positions 68120-68810 (Figure 2B).
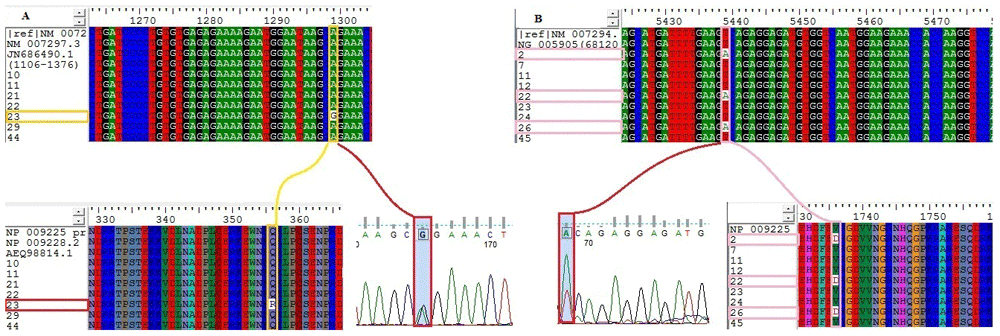
(A) Heterozygous substitution from Glutamine (Q) to Arginine (R) at position 356 in patient 23, due to missense substitution mutation from Adenine (A) to Guanine (G) c.1299A>G, coding sequence. (B) Heterozygous substitution from Valine (V) to Aspartic acid (D) at position 1736 in patients 2, 22 and 26 due to missense substitution mutation from thymine (T) to adenine (A) c.5439 T>A coding sequence. Using the transcript variant 1 NM_007294 mRNA, which represents the complete BRCA1 coding sequence used to align all patient sequences under screening, and the corresponding amino acid sequences NP_009225 to align all patient translated amino acid sequences.
SNP information. SNP information [SNP ID, MIM: 113705, RefSeq Gene accession No.: NG_005905 on chromosome 17, mRNA accession NO.: NM_007294 transcript variant 1 with 7224 bp and Protein accession numbers : (NP_009225) protein isoform 1 with 1863 a.a. and P38398 UniProt entries] concerning the human BRCA1 gene, which was used in our computational analysis, was retrieved from the NCBI database of SNPs: dbSNP (https://www.ncbi.nlm.nih.gov/snp).
SNP prediction. SNPs were analyzed using five prediction online tools: SIFT (http://sift.bii.a-star.edu.sg/)56, Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/)57, I-MutantDDG-Seq Suite (http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi) and PhD-SNP (http://snps.biofold.org/phd-snp/phd-snp.html)58. The tertiary model of protein and mutation analysis was done online using Hope software (http://www.cmbi.ru.nl/hope/input)59. MutationTaster2 (http://www.mutationtaster.org/) was used to assess the protein features of the resulting variants, with comprehensive (input/output) criteria, which predicts potential disease-causing mutations60.
Two missense variants were detected within the study, one patient with Q356R and three patients with V1736D. Both variants were heterozygous (Figure 2) and were detected within premenopausal patients, with a mean age of 37 years. Three patients were multiparous; the one case of (Q356R) and two cases of (V1736D) were multiparous (mean parity, 2.8). There was no family history of BC in patients with the two variants (Table 3).
The patients’ sequences, and the additional nucleotide sequences with the gene sequence, were aligned against the reference standard sequence in the RefSeqGene and nucleotide databases (accession, NM_007294), which is BRCA1 gene transcript variant 1, and was introduced to replace the previously existing sequence (gi: 63252871) in May 200961. Two substitutions-bearing monoallelic alterations were found, the first one at position 1299 (A/G) located in exon 11 (Figure 2), and the second one at position 5439 (T/A) in exon 20 (Figure 2). Primer 1 has been excluded from the study because of the errors that have been noticed within all patient sequence data chromatogram results.
After translation to amino acid sequences, the samples were aligned against BRCA1 protein isoform 1 (accession, NP_009225). Q356R was found to meet its corresponding nucleotide change and position c.1299A>G in which Glutamine (Gln) replaced by Arginine (Arg) in patient 23, and V1736D was found to meet its corresponding nucleotide change and position c.5439T>A, in which Valine (Val) was replaced by Aspartic acid (Asp) in patients 2, 22 and 26 (Figure 2). These variants (Q356R, V1736D) were then predicted with SIFT, Polyphen-2, I-Mutant-3, PhD-SNP and MutationTaster2 software to obtain their pathological effects, and results are provided in Table 4. Amino acid properties for the wild Val and the mutant Asp residues and the 3D structure of the variant V1736D were obtained using Project Hope software (Figure 3A).
SNP: single nucleotide polymorphism; RI: reliability; DDG: ΔΔG; SVM: support vector; SVM2 value: DDG < 0: decrease stability, DDG >0 increase stability machine; DDG value: DG (New Protein)-D (Wild Type) in Kcal/mole.
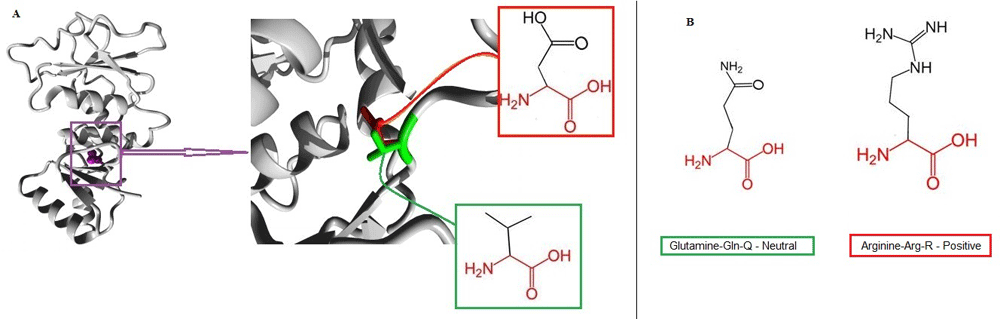
(A) This illustration shows the BRCA1 protein in grey colour on the (left) with the magenta coloured side chain of the mutated residue, and on the (right) the higher magnification of the area of mutation displays the side chains of both amino acid residues more clearly, the green highlights the wild type Valine, and the red highlights the mutant type Aspartic acid with their schematic structures, as both contain the same backbone (red) but they differ in their side chains (black). (B) The illustration shows the schematic structural units of the neutral wild type Glutamine (left) and the positive mutant Arginine (right). Both amino acids contain the same backbone (Red) but they differ in their side chains (Black).
In this study, we found two mutations-bearing monoallelic features. One missense mutation rs1799950 in exon 11 BRCA1 gene in patient 23: position number 1299 (A/G) as (CAGA) to (CGGA), using transcript variant 1 (NM_007294), which led to change in the coding amino acid (Glutamine-Gln-Q) to (Arginine-Arg-R) at position 356 using protein isoform 1 (NP_009225.1) (Figure 2A).
The other was a novel SNP, situated at exon 20, with higher frequency in patients 2, 22 and 26: position 5439 (T/A) as (GTCA) to (GACA), which led to change in the coding amino acid (Valine-Val-V) to (Aspartic acid-Asp-D) at position 1736 (Figure 2B). Both SNPs (Q356R) and (V1736D) were found to affect the resulting translated protein. According to Project Hope software, starting with the novel (V1736D) variant, we found that the mutant residue Aspartic acid (D) of the outcome BRCA1 mutated protein is located near a highly conserved position. The mutant residue is bigger and less hydrophobic than the wild-type residue. The wild-type residue was neutral, the mutant residue is negatively charged. The mutated residue is located in a domain that is important for binding of other molecules, as it is in contact with residues in another domain. It is possible that the mutation can disturb these contacts. It may also disturb the interaction between these two domains, which would affect the function of the protein; therefore, it might disturb signal transfer from binding domain to the active domain. This novel mutation was detected in patients from different tribes from Sudan, who were all ≤45 years old.
In the case of the Q356R mutation, the mutant residue of Arg is positively charged compared to the neutral wild type Gln residue. This could lead to the repulsion of ligands or other similar charged residues. The size of the end variant is bigger than the wild-type residue and this might lead to bumps, as reported by Project Hope software (Figure 3B). In addition, the mutated residue is located on a domain responsible for protein activity; hence the activity could be altered by its physical variation conferred by its new charge and size. The Arg 356 variant does, however, generate a run of three positively charged residues (Lys Arg Lys), and as a result it could alter the properties of the protein, which is composed of 16% negatively charged residues overall62. In the current study, we found the same result when it was described as a disease-related mutation of altered protein stability (Table 4).
According to MutationTaster2, the novel variant (V1736D) was predicted to be ‘disease-causing’, and the software assessed and calculated the probability of such variation on the resulting protein, and showed that the new features would be disease-causing and displayed it with a higher score. By contrast, the software predicted that in the case of Q356R, the resulting Arg was a polymorphism of little harm, and reported it with a lower score compared to the protein feature of the other variant, and reports from the other softwares used for assessment (Table 4). Q356R has not yet been classified in terms of clinical significance in NCBI dbSNP.
Two novel variants were identified to be deleterious within the BRCA1 gene among premenopausal women patients in two local studies9,35. One variant was a truncated stop codon and the other was predicted computationally: c.3999delT, stop codon 1335, and c.5090G>A, p.Cys1697Tyr, respectively. In addition to these variants, a deleterious mutation, c.4986+6T>C, located in intron-exon boundary was found in the youngest patient of 25 years in one of the studies35. These findings, compared to our present study finding (V1736D), showed that early onset BC is associated with a deleterious nature of the identified variants. In addition, our study included pre and postmenopausal patients, all detected variants were mainly confined to premenopausal cases. Therefore, genetic studies are highly recommended to highlight the genetically susceptible nature of patients diagnosed with early onset disease, who may harbor the deleterious variants that could have a significant role in developing BC. Two patients within the present study were screened previously in a local study targeting pathological SNPs within BRCA2 gene selected regions, which identified a stop codon (L1053X)63. One of these patients was the youngest patient identified with Q356R, and the other presented with the bilateral disease and identified with V1736D. Both patients were reported to have stop codons at position L1053X with nucleotide and protein sequences identifiers of KT901810 and ALQ44030, and KT901807 and ALQ44027, respectively63.
The SNP found in the present study, c.1299A>G, matched a previously reported SNP, in the same altered nucleotide position 1299A>G, and the same altered outcome protein position, Q356R42,64. The same protein position (Q356R) with the same altered nucleotide (A/G) has also been described previously, but with a different nucleotide position c.1186 A>G16,65,66. Some studies found that this SNP is associated with patients under 40 years old,66,67, which agreed with our study - the youngest premenopausal patient (27 years old) was detected with this variant. Although this variant’s effect remains uncertain, a previous finding to our study has described the same mutation, Q356R in Moroccan BC patients, which was the first study that described this mutation in a North African population66. They found that most previous studies that described this mutation were within western European populations68 and there were no studies found in a North African population to confirm this, thus more research is needed to investigate this. Our study is closer to the Moroccan study regarding the early onset characteristic this variant has, revealed by the similarity in the background history of the patient with a Q356R polymorphism in our study and those detected in the Moroccan study, who also did not have a family history of BC66. This variant has been reported to be independently minor or leads to a very slightly increased BC risk, but a risk that is cumulatively significant69. In another study, this mutation was found in patients with a family history of ovarian cancer, suggesting that this variant may increase ovarian cancer risk65,70. Both detected variants in the present study were identified to have a pathological effect, with the exception of the results from MutationTaster2 in the case of Q356R. Therefore, the Q356R mutation may have a pathological effect, and the novel SNP might play a role in the pathogenesis of the disease.
In order to only have clear DNA sequence results for comparison with NCBI references, primer 1 results were excluded due to sequencing errors (see Supplementary File 1). In addition, three patient sequences of primer 2 set and one patient of primer 3 set have been excluded for the same sequence errors.
The limitation of this study was the small sample size and the functional assessment facilities available to assess the protein of monoallelic alteration for their pathological contribution to the disease. Financial constraints also limited the study. Therefore, we recommend further studies in a larger number of Sudanese patients to further explore these findings.
In the present study, Sudanese BC patients were investigated for BRCA1 mutations. Two different mutations were found in young patients of ≤45 years old, with no family history of BC. To conclude, the study has highlighted a need for further research of these mutations amongst a larger population (including patients and controls), so as to investigate the variants’ distribution through the population and their potential diagnostic value. This will aid the understanding of a variant’s frequency and clinical significance. In addition, both variants identified, require in vitro functional and protein level assessment.
The BRCA1 sequence data of the novel variant (V1736D) from this study has been submitted to NCBI GenBank under the accession numbers and protein identifiers found in Table S1.
Dataset 1: BRCA1 sequence results in a zipped file. These sequencing results as received from Macrogen Company (Seoul, South Korea) comprise all the breast cancer patients in this study using the three sets of primers (1, 2 and 3).Each patient has the sequencing data in different file formats (a sequencing data file that needs to be viewed by a sequencing viewer software, i.e FinchTV; a PDF; and a FASTA format text document). doi, 10.5256/f1000research.11395.d17244571
Dataset 2: Patient demographics (non-identifying) according to primer. Patient demographics, clinical data, and histological parameters with highlighted detected missense variants (primers 2 and 3) of each 10 selected subset patients. doi, 10.5256/f1000research.11395.d17244672
MEMMA, AAME, MAS conceived the study. ME-FME, HNA, MMAH, MAS designed the experiment. MEMMA, AAME, sample collection and lab preparation. ME-FME, MMAH designed the wet lab practice. MEMMA, AAME, ME-FME practiced the wet lab methodology. MAS, HNA, AAME, MMG, AAF, MMO, SAO, HAS, MSA, TS, RAO, RAE bioinformatics assessment. AAME, MMG, AAF, MMO, SAO, MSA, TS prepared the manuscript draft. AAME, MMG, MSA, MMAH, MAS revised and edited the manuscript draft. MMAH revised the final edited manuscript.
The authors extend their heartfelt thanks to Prof. Mohamed Ahmed Salih for his priceless help and palatable advice, Al-Neelain University-Khartoum state, Radiation and Isotope center- Khartoum state and Africa city of technology-Khartoum state members for their help. And we would also love to thank Ms. Catherine Russel and Mr. Omar Awad Elimam for their kind support in language corrections.
Supplementary File 1: Evidence for sequencing errors of primer 1. The file contains FinchTV snapshots, and the uncropped western blot for primer 1.
Click here to access the data.
Table S1: NCBI GenBank accession numbers for the novel BRCA1 sequence data from this study.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Borg A, Haile RW, Malone KE, Capanu M, et al.: Characterization of BRCA1 and BRCA2 deleterious mutations and variants of unknown clinical significance in unilateral and bilateral breast cancer: the WECARE study.Hum Mutat. 2010; 31 (3): E1200-40 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Enzymes and cell signalling, protein expression, gene expression
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Enzymes and cell signalling, Protein expression, gene expression.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genetics, molecular genetics, cytogenetics, genetic counseling
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 4 (revision) 24 Jan 18 |
read | ||
|
Version 3 (revision) 06 Nov 17 |
read | ||
|
Version 2 (revision) 21 Sep 17 |
read | read | |
|
Version 1 14 Aug 17 |
read | read | |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)