Keywords
Biological screening data, statistical analysis, active compounds, assay frequency, hit rate distribution, assay interference
This article is included in the Cheminformatics gateway.
Biological screening data, statistical analysis, active compounds, assay frequency, hit rate distribution, assay interference
Compounds with false-positive signals in biological assays cause substantial problems for biological screening and medicinal chemistry1. Assay artifacts often remain undetected or are unveiled only at later stages of compound development efforts, leading to substantial loss of time and resources. Moreover, once published, artificial activities spread through the scientific literature and potentially cause even more harm by inspiring follow-up investigations that are doomed to fail. Known assay interference compounds include colloidal aggregators2–7 and many other compound classes that can react in different ways or are fluorescent under assay conditions6–15. Systematic efforts to identify interference compounds include the compilation of aggregators2–4 and pan-assay interference compounds (PAINS)8,9. The latter comprise a set of 480 classes of compounds originally identified in AlphaScreen assays8. PAINS are typically contained as substructures in larger compounds. However, the assessment and prediction of assay interference is far from being a trivial exercise. For example, analysis of screening data from PubChem16 has revealed that many compounds containing PAINS, including most reactive chemical entities, have very different hit rates or might be consistently inactive17,18. Moreover, analogs or different series of analogs containing the same PAINS substructure often have distinct activity profiles and are active against different targets19. Thus, interference characteristics of related compounds frequently differ and a substructure with interference potential does not necessarily give rise to false-positive assay signals. To further complicate matters, promiscuous compounds may also have true multi-target activities20 that are relevant for polypharmacology20–22. Moreover, even highly promiscuous screening hits include molecules with no apparent liabilities, in addition to obvious interference compounds12.
Without doubt, judging assay interference and candidate compounds requires profound chemical knowledge and experience. It is equally relevant, however, to strive for a data-driven assessment of promiscuity by exploring compound activity data on a large scale20, aiming to identify compounds with interference potential for further analysis. Therefore, we have carried out a statistical analysis of hit rates of compounds that were extensively tested in screening assays. A special feature of this study has been its focus on pairs or larger series of analogs, rather than single compounds, which provides additional confidence criteria for activity assessment and further increases the information content of activity data analysis. Many series of analogs with much higher than typically observed hit rates and largely consistent activity profiles across many different assays were identified. This collection of series provides a basis for further investigating compounds with interference potential or true multi-target activities.
All calculations were carried out using in-house scripts and implementations.
From the PubChem BioAssay database16, 437,257 compounds were pre-selected that were tested in both primary and confirmatory assays, representing extensively assayed screening compounds23. Approximately 95% of these compounds were evaluated in more than 50 primary and/or confirmatory assays23. Primary PubChem assays report compound activity (e.g., percentage activity) for a single dose, while confirmatory assays are dose-response assays yielding titration curves and IC50 values. Our current analysis focused on primary assays, for which much larger data volumes were available than for confirmatory assay. Primary assays also included assays for which no target was specified (such as cell-based assays). For pre-selected compounds, hit rate statistics were determined.
A matched molecular pair (MMP) is a pair of compounds that are only distinguished by a chemical change at a single site24, termed a chemical transformation25. As an extension of the MMP concept, a matched molecular series (MMS) was defined as the union of all MMP compounds that are only distinguished by chemical modifications at a given site26. Accordingly, an MMS represents a series of analogs sharing a single substitution site. To generate MMPs, exocyclic single bonds in screening compounds were systematically fragmented25 following retrosynthetic fragmentation rules27, yielding so-called RECAP-MMPs28. These MMPs were subject to transformation size restrictions in order to limit chemical changes to modifications typically observed in series of analogs29. An MMS was designated as redundant if it was a subset of a larger MMS or if there was another MMS representing the same series of analogs but having a larger MMP core. For screening compounds with high hit rates, non-redundant MMS were systematically determined.
For each MMS, three parameters were calculated. First, the MMS hit rate (HR) was obtained from the union of all assays (i.e., the number of unique assays in which one or more analogs were tested in) and assays with activity signals (active assays, i.e., the number of unique assays in which one or more analogs were found to be active). Second, assay overlap was determined as the proportion of assays in which all MMS compounds were tested in (shared assays, i.e., the intersection of assays) relative to the union of assays. Third, from assay overlap, assays with inconsistent activity were calculated as the proportion of shared assays in which different MMS compounds were active or inactive.
A statistical analysis of hit rates of extensively tested screening compounds is presented taking assay frequency into account. On the basis of the hit rate distribution, ranges of unusually high hit rates were determined. From compounds with high hit rates, analog series with single substitution sites (MMSs), i.e., “minimal” chemical modifications within series, were systematically extracted, which provided structural context information and hit rate controls for closely related compounds. For MMSs, different parameters were calculated, making it possible to compare and prioritize these series. The collection of MMSs with high hit rates provides a basis for investigating assay interference candidates, as well as chemical entities with potential multi-target activities.
Figure 1 (boxplot on the left) shows the global distribution of primary assays for 437,257 extensively tested PubChem compounds, with a median value of 347 assays per compound. From these, a subset of 327,532 compounds was selected that were tested in more than 257 primary assays, corresponding to the lower quartile boundary of the global distribution. For this subset, the assay distribution was separately monitored (Figure 1, boxplot on the right), yielding a median of 426 (and a maximum of 626) assays per compound. Hence, half of these compounds were tested in more than 426 primary assays.
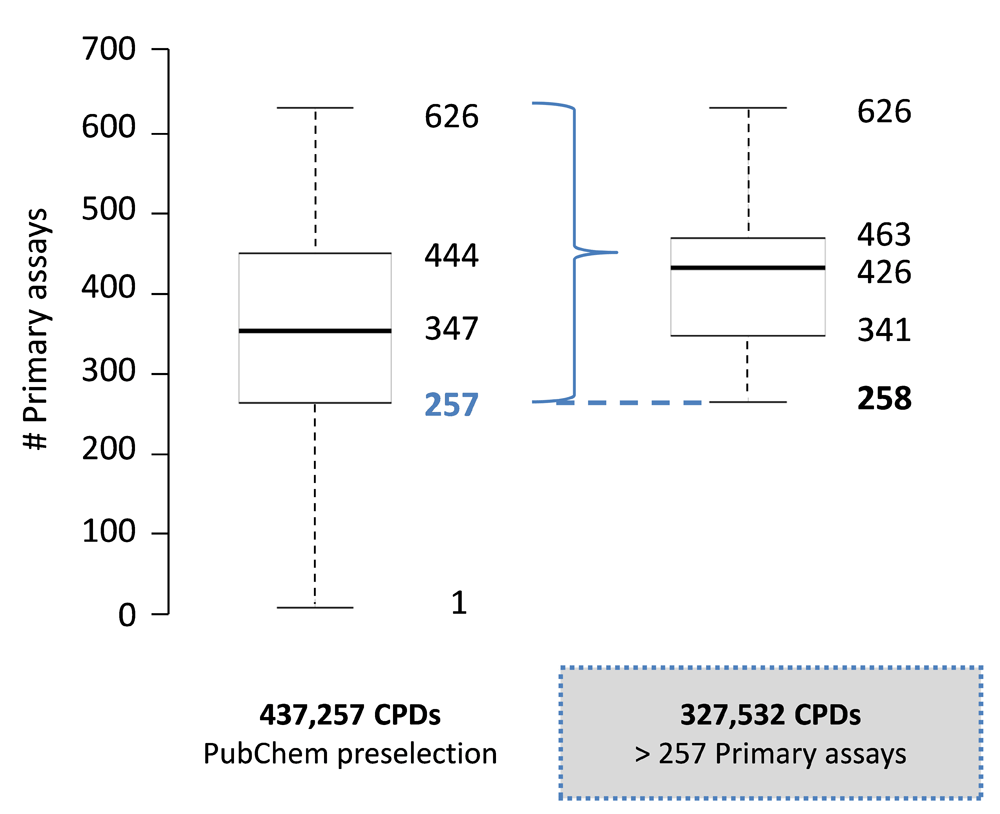
The frequency distribution of primary assays is shown in a boxplot format for 437,257 pre-selected PubChem compounds and a subset of 327,532 compounds. The plot gives the smallest number of primary assays (lower whisker), first quartile (lower boundary of the box), median value (thick line), third quartile (upper boundary of the box), and largest number of assays (upper whisker). Outliers are not displayed. The dashed blue line indicates the selection criterion for the compound subset (i.e., tested in more than 257 primary assays).
For 327,532 compounds tested in more than 257 assays, hit rates were determined. The distribution is reported in Figure 2 (boxplot on the left), resulting in a median hit rate of 0.4%. The lower quartile boundary and lower whisker of the boxplot were identical and represented consistently inactive compounds, which were not of interest for our current analysis. On the basis of the distribution, the interval of “bulk hit rates” (b_hr) for these extensively assayed PubChem compounds was defined as 0% < b_hr ≤ 1.0%, covering the lower quartile, median, and upper quartile (and hence the “bulk” of the distribution). There were 80,495 compounds with hit rates ≥ 1.0%. The hit rate distribution of this compound subset is shown in Figure 2 (middle), yielding a median of 1.8%. This value was set as the hit rate threshold for most active screening compounds. The threshold was exceeded by 41,609 compounds, representing 12.7% of the initial compound pool. The hit rate distribution of these compounds is reported in Figure 2 (right), resulting in a median of 2.9%. We determined that 93.1% of the compounds with hit rates greater than 1.8% in primary assays were also active in confirmatory assays (yielding IC50 values). Hence, their activity was not confined to primary assays.
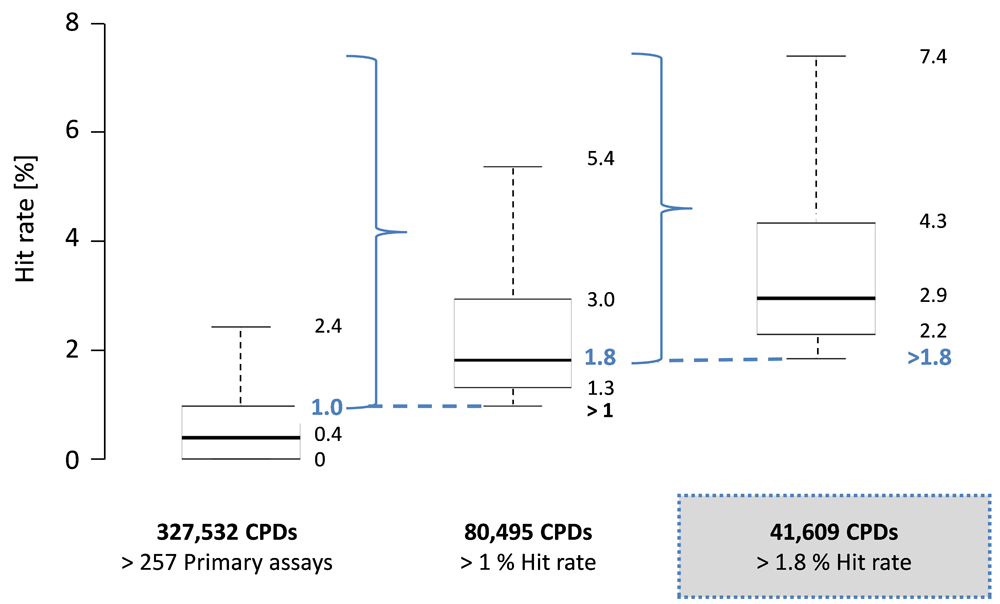
For three different subsets of PubChem compounds, hit rate distributions are shown in boxplots according to Figure 1. The subsets are characterized by increasing hit rates (marked by dashed blue lines).
From the 41,609 compounds with highest hit rates, MMSs were systematically extracted on the basis of RECAP-MMPs. After removal of redundant MMSs (see Methods), 6941 unique MMSs were obtained comprising 14,646 compounds, which represented our final hit rate- and series-based selection set. Table 1 reports the size distribution of the MMSs, ranging from two to 17 analogs per series. With 6111 instances, compound pairs and triplets dominated the distribution, but more than 800 larger MMSs were also obtained. As further discussed below, compound pairs and triplets already provide informative controls for activity analysis and enable a more confident assessment compared to the analysis of individual compounds. This was a major motivation for focusing the analysis on MMSs.
The distribution of 6941 frequently active MMSs (#MMSs) over increasing numbers of compounds (#CPDs) is reported.
| #CPDs | #MMSs |
|---|---|
| 2 | 4965 |
| 3 | 1156 |
| 4 | 435 |
| 5 | 190 |
| 6 | 70 |
| 7 | 48 |
| 8 | 22 |
| 9 | 21 |
| 10 | 11 |
| 11 | 12 |
| 12 | 3 |
| 13 | 4 |
| 14 | 2 |
| 15 | 1 |
| 17 | 1 |
Figure 3a illustrates the derivation of three parameters for the characterization and comparison of MMSs (rationalized in the Methods section). The cumulative MMS hit rate is a direct measure for the activity of a series. In addition, assay overlap represents a confidence criterion for MMS assessment, i.e., large assay overlap of compounds comprising a series assigns high confidence to hit rate comparisons. By contrast, the proportion of assays with inconsistent activity should best be minimal to draw firm conclusions. Figure 3b reports the distribution of these three parameters for the 6941 MMSs. Assay overlap (upper left plot) and MMS hit rates (lower left) were generally high, with median values of 79.3% and 5.8%, respectively. By contrast, the proportion of inconsistent assays (upper right) was overall low, with a median of only 3.7%. Thus, the distributions of MMS parameters indicated that the set of MMSs was suitable for the analysis of series-based hit rates and hit rate comparison of compounds comprising individual MMSs. We note that MMSs can be ranked in the order of decreasing assay overlap and MMS hit rates and increasing inconsistent assays and prioritized, for example, on the basis of rank fusion calculations.
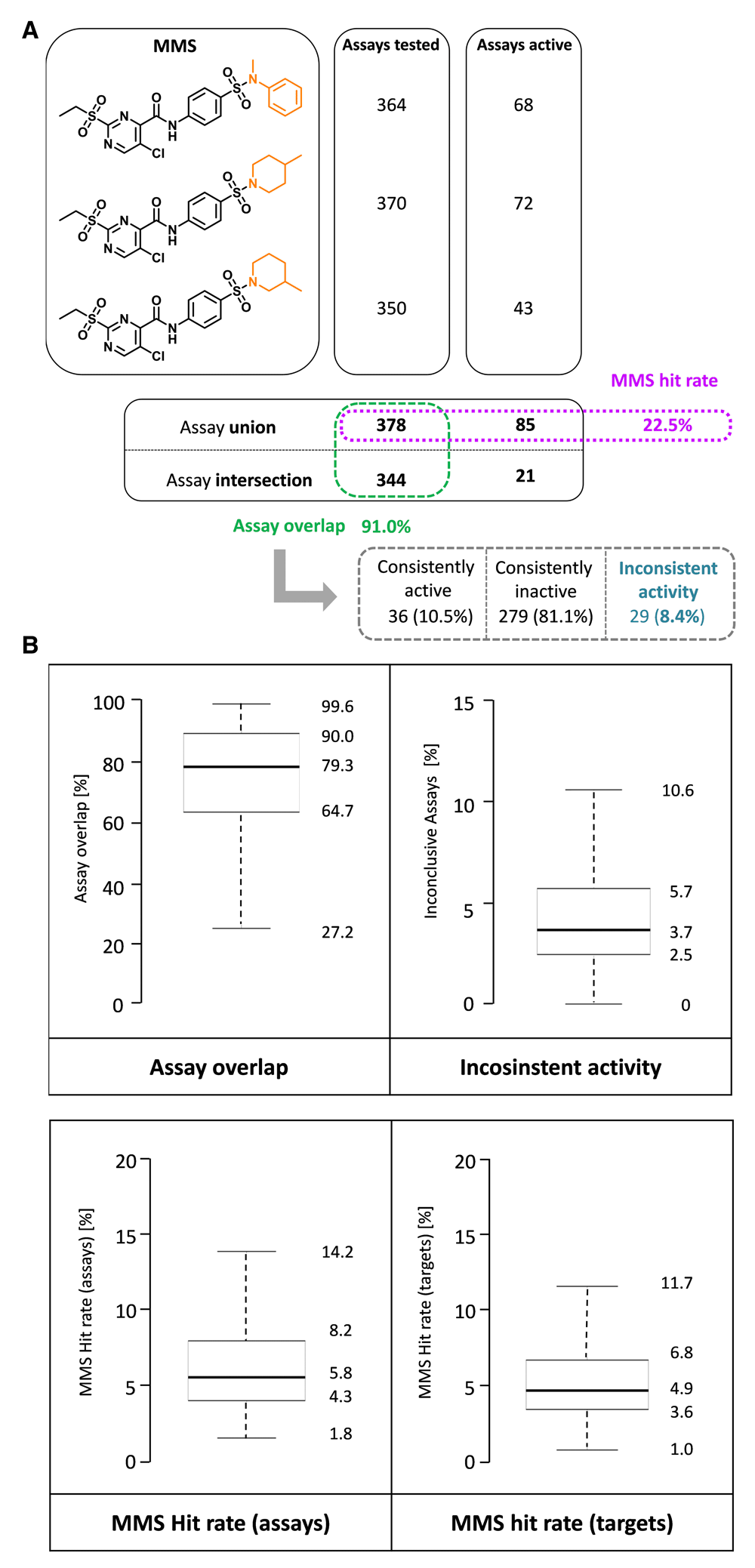
(a) An exemplary MMS comprising three analogs is shown. The MMS core and varying substituents are colored in black and orange, respectively. For each compound, the number of assays it was tested and active in is reported, respectively. Furthermore, the assay union, intersection, and MMS hit rate (purple) are given. From these data, the assay overlap (green) of MMS analogs was determined as well as the proportion of assays with consistent activity, inactivity, and inconsistent activity (blue). (b) Boxplots are shown reporting the distribution of assay overlap, assays with inconsistent activity as well as assay- or target-based MMS hit rates for PubChem compounds with greater than 1.8% hit rate.
Our analysis was intentionally focused on hit rates over assays (i.e., assay promiscuity) to take as many activity readouts as possible into account. Therefore, as a control, assay- and target-based hit rates were also compared. Compounds forming the 6941 MMS were evaluated in a total of 1213 assays. For 255 of these assays, no individual target was specified. The remaining 958 assays covered 426 different targets. Figure 3b reports the distributions of MMS hit rates over assays (lower left plot) and targets (lower right). The distributions were overall similar, with median values of assay- and target-based hit rates of 5.8% and 4.9%, respectively. Hence, despite the presence of multiple assays for a subset of targets, assay-based hit rates were only slightly higher than target-based rates, indicating that corresponding conclusions would be drawn from the analysis of these distributions.
The computational aggregation advisor4 and compound strings taken from PAINS filters30,31 (http://www.rdkit.org) were used to search the MMSs for known assay interference candidates. The 14,646 MMS compounds contained 783 aggregators (on the basis of 100% similarity) and 2381 compounds with PAINS substructures. There were 611 MMSs with one or more aggregators, 1139 MMSs with one or more PAINS, and 126 MMSs including aggregators and PAINS. However, 5065 MMSs with high hit rates did not contain known compounds with aggregation potential or PAINS substructures. Thus, the MMSs provide a large source of analogs for the exploration of other interference candidates, as well as compounds with true multi-assay/target activities.
Figure 4 shows exemplary compound pairs and triplets with high assay promiscuity. The two analogs in Figure 4a were tested in more than 380 assays with 93.5% assay overlap and only 1.6% inconsistent assays, yielding comparable hit rates of 2.8% and 3.2%, respectively, resulting in an MMS hit rate of 4.5%. These analogs contained a classical PAINS substructure (ene_rhodanine)8,9. Furthermore, compounds in Figure 4b were analogs of a molecule with aggregation potential4. They were tested in more than 300 and 400 assays, respectively, yielding a relatively low assay overlap of 59%, and had hit rates of 2.2% and 2.6%, respectively, resulting in a low MMS hit rate of 2.9%. Thus, these analogs were far from being consistently active, as one might assume for strong aggregators. In Figure 4c, a pair of thieno[2,3-d]pyrimidine-2-acetic acid ethyl ester analogs is shown that were tested in 442 assays with large overlap. These compounds had high hit rates of 5.9% and 7.9%, respectively, resulting in a high MMS hit rate of 9.8%. Moreover, Figure 4d shows a triplet of sulfonylpyrimidines that were tested in 357–361 assays with 89.7% overlap, having very high hit rates of 8.7% (one analog) and more than 13% (two analogs). Compounds forming each of the MMSs in Figure 4 displayed consistent hit rate characteristics, hence assigning confidence to their observed activity phenotype. The analogs in Figure 4c and Figure 4d have previously not been classified as interference candidates. However, they might well be reactive under assay conditions. Taken together, these examples of analog pairs and triplets (i.e., minimally sized MMSs) are indicative of the potential of well characterized MMSs for follow-up investigations focusing on assay interference and multi-target activities.
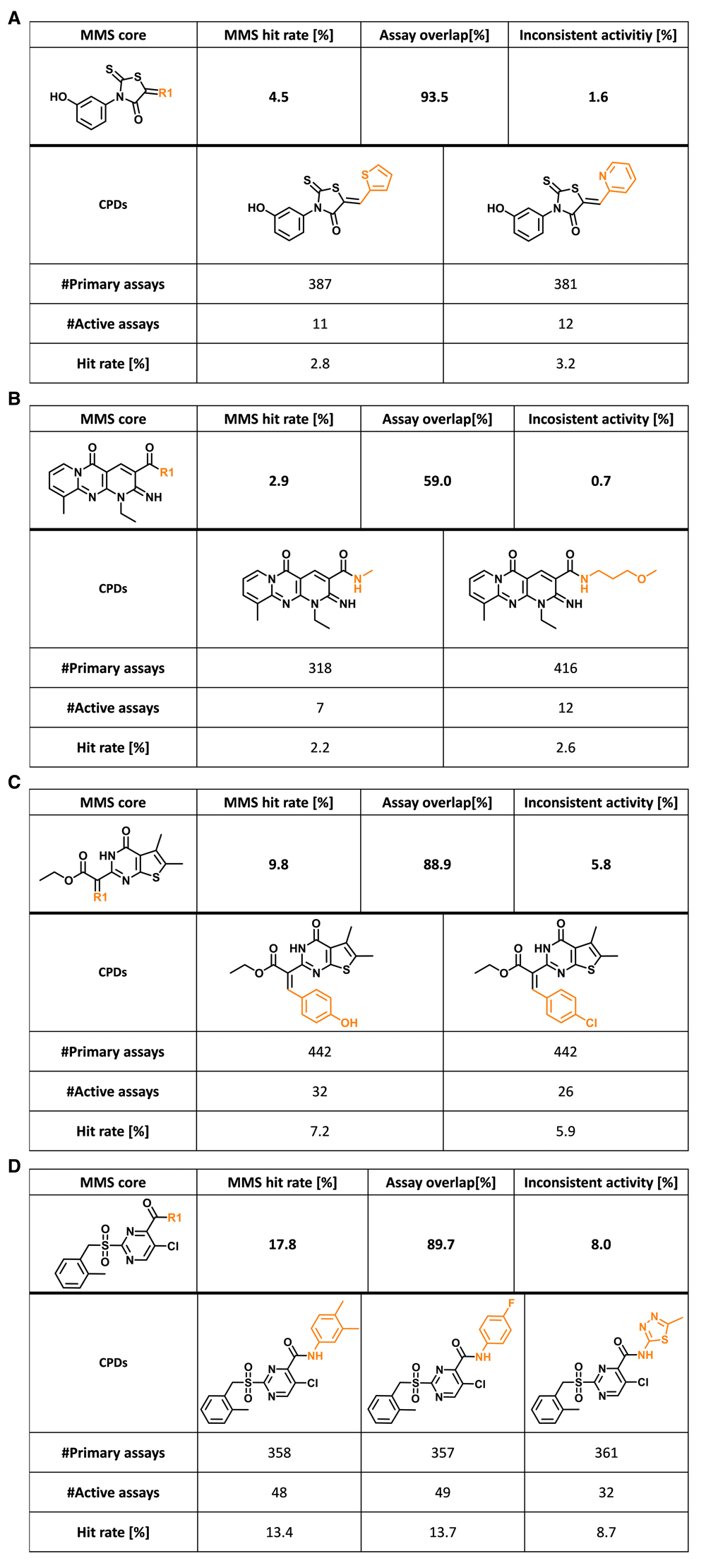
(a–d) Four exemplary MMSs (core, black; substituents, orange) are shown and the MMS hit rate, assay overlap, and proportion of assays with inconsistent activity are reported. In addition, for each individual analog, its assay frequency and hit rate are provided.
Herein, a detailed analysis of hit rates of nearly 440,000 extensively assayed screening compounds has been presented. On the basis of hit rate distributions, 12.7% of the compounds with highest hit rates were selected. From these compounds, analog series with single substitution sites were systematically extracted to complement hit rate statistics with the assessment of structural relationships between active compounds. A total of 6941 unique MMSs were obtained comprising 14,646 compounds. These MMSs were characterized using different parameters prioritizing high-confidence series for activity analysis. A major goal of our study has been the data-driven generation of a pool of analog series for the evaluation of assay interference potential and multi-target activities. More than 5000 MMSs did not contain known interference candidates, providing an opportunity to evaluate compounds with interference potential on a large scale. In the next step, analog series will be evaluated from a medicinal chemistry perspective to complement and further extend statistical considerations. Annotated series and associated assay/target information will then be made freely available. The statistics and selection steps reported herein also make it possible to regenerate compound subsets at different hit rate levels and subject them to further analysis. In addition, large numbers of compounds with high hit rates that were not part of MMSs are also available. For reasons discussed, our preferred approach is taking compound series information into account when judging assay promiscuity.
The data sets used in this study are freely available in PubChem and can be generated following the selection protocol reported in the Methods.
DS is supported by Sonderforschungsbereich 704 of the Deutsche Forschungsgemeinschaft.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: assay interference, medicinal chemistry
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Hu Y, Bajorath J: Entering the 'big data' era in medicinal chemistry: molecular promiscuity analysis revisited.Future Sci OA. 2017; 3 (2): FSO179 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 09 Oct 17 |
read | read | |
|
Version 1 17 Aug 17 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
On a more general note, although we are among the computational groups promoting open science by public release of many in-house generated data sets and software tools, our experiences with free data and tool sharing have not been entirely positive; another point of consideration going forward. Perhaps it might make sense to (re-)consider other collaborative models and release procedures.
On a more general note, although we are among the computational groups promoting open science by public release of many in-house generated data sets and software tools, our experiences with free data and tool sharing have not been entirely positive; another point of consideration going forward. Perhaps it might make sense to (re-)consider other collaborative models and release procedures.