Introduction
α-arrestins are a family of proteins that contain arrestin domains whose sequences and structures have similarities with those of classical visual and β-arrestins1–3. α-arrestins are considered as ancestral forms of arrestins because their orthologs exist in fungi, including yeast, which do not have visual or β-arrestins1,4. Several mammalian α-arrestins modulate metabolism and receptor desensitization5,6, but much remains to be elucidated concerning the functions of α-arrestins in many organisms. The human genome encodes 6 α-arrestins and 4 visual or β-arrestins. Interestingly, the simple roundworm Caenorhabditis elegans genome contains 29 α-arrestin and 1 β-arrestin genes1,7. Therefore, the C. elegans system provides opportunities for the genetic analysis of α-arrestins in various aspects of physiology both individually and combinatorially. However, information regarding the functions of C. elegans α-arrestins is limited8,9.
C. elegans is an excellent genetic model organism that has been exploited for studying conserved biological processes, including apoptosis, behavior, development and aging. In particular, its short lifespan in combination with genetic amenability has made C. elegans one of the most popular models for research on aging and longevity10,11. Many factors, including components in insulin/insulin-like growth factor 1 (IGF-1) signaling (IIS), have been identified as lifespan and aging regulators in C. elegans11–13. For example, genetic inhibition of IIS components, such as DAF-2/insulin/IGF-1 receptor, robustly extends lifespan and delays physiological aging through up-regulating transcription factors, including DAF-16/FOXO11–13. Importantly, the roles of these aging-regulatory factors in C. elegans have been shown to be conserved in other species, including Drosophila and mammals12,13. One of the powerful ways to identify novel factors that influence aging is by employing genetic screens, such as an RNA interference (RNAi) screen. We previously identified several genetic factors, including RNA helicases, that modulate longevity in C. elegans, through targeted or genome-wide RNAi screens14–17. Because of its robust longevity phenotype that confers sensitivity to changes in lifespan, daf-2/insulin/IGF-1 receptor mutants serve as an excellent platform for the identification of novel lifespan-regulating factors17,18.
In this study, we aimed to determine whether any α-arrestins played a role in the lifespan regulation of wild-type or daf-2 mutants. We performed a lifespan assay-based RNAi screen targeting 24 out of 29 C. elegans α-arrestins. We found that knocking down each of the α-arrestins had small or no effects on the lifespan of wild-type and daf-2 mutants. Thus, C. elegans α-arrestins may play minor or modulatory roles in lifespan regulation. Based on our results, it will be important to test the roles of α-arrestins in combinatorial manners and/or by using strong loss-of-function mutations in future research.
Methods
Caenorhabditis elegans strains
All strains were maintained as previously described19. The following strains were used in this study: N2 wild-type, CF1041 daf-2(e1370) III outcrossed six times to N2.
Phylogenetic analysis
The protein sequences of 27 α-arrestins, except arrd (arrestin domain protein)-20 and arrd-21, were obtained from Wormbase (www.wormbase.org, version WS259). The protein sequences of arrd-20 and arrd-21 were obtained from Ensembl (http://www.ensembl.org, release 89). The phylogenetic tree of 29 α-arrestins in C. elegans was generated using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/)20 and re-visualized using the Dendroscope 3 (version 3.5.9)21. For the α-arrestins that have multiple isoforms, isoform a was used for the analysis.
RNAi clones
Twenty one RNAi clones that target C. elegans α-arrestin genes were used from two commercial C. elegans RNAi feeding libraries. Specifically, RNAi clones targeting arrd-2, arrd-6, arrd-7, arrd-8, arrd-9, arrd-10, arrd-13, arrd-16, arrd-18, arrd-23, arrd-24, arrd-25, arrd-28 and ttm-2 (toxin-regulated targets of MAPK 2) were obtained from Ahringer laboratory library (Geneservice Ltd., Cambridge, UK), while those against arrd-1, arrd-3, arrd-4, arrd-5, arrd-14, arrd-15 and arrd-19 were from Vidal laboratory library (Source BioScience, Nottingham, UK). Three RNAi clones targeting arrd-11, arrd-17, and arrd-26 were generated by molecular cloning using infusion recombinase (EZ-FusionTM Cloning Kit, Enzynomics, Daejeon, South Korea). The N2 genomic DNA was obtained through the lysis of worms using proteinase K (Invitrogen, Carlsbad, CA, USA) and N2 complementary DNA was obtained from RNA extraction using RNAiso Plus (Takara, Shiga, Japan) followed by reverse transcription using ImProm-II reverse transcriptase (Promega, Madison, WI, USA, USA). The infusion reaction between PCR products and pL4440 plasmids (Fire lab C. elegans vector kit, 1999) digested with HindIII (New England Biolabs, Ipswich, MA, USA) and Acc65I (New England Biolabs) was followed by transformation of in-house competent E. coli HT115 cells and by selection of positive clones on ampicillin (USB, Santa Clara, CA, USA)-containing LB plates. Primers (CosmoGenetech, Seoul, South Korea) that were used to amplify coding regions of arrd-11 from N2 genomic DNA, and those of arrd-17 and arrd-26 from N2 complementary DNA are as follows: forward primer 5’-GAATTCGATATCAAGCTCCCTCGTGCAAATTAGGAAA-3’ and reverse primer 5’-CTATAGGGCGAATTGGGGTTCCTCCCACTCCATACA-3’ for arrd-11; forward primer 5’-GAATTCGATATCAAGCTATGGTGCAGTTAGATCGTTTTG-3’ and reverse primer 5’-CTATAGGGCGAATTGGTTAATCGGTATAAAATGG-3’ for arrd-17; forward primer 5’-GAATTCGATATCAAGCTATGAAGGTCGATTACTTCG-3’ and reverse primer 5’- CTATAGGGCGAATTGGCTACTTCTCGGAGCCATTTG-3’ for arrd-26. The sequences of all these 24 α-arrestin RNAi clones were confirmed using DNA sequencing (Solgent, Daejeon, South Korea) before lifespan assays. RNAi clones targeting remaining five α-arrestin genes were not generated.
RNAi screen using lifespan assay
The RNAi screen employing lifespan assay was performed as previously described17. Briefly, E. coli HT115 bacteria that expressed specific double-stranded RNAs (dsRNAs) were cultured overnight at 37°C in LB media containing 50 μg/ml ampicillin (USB). The cultured bacteria were seeded onto nematode growth media plates containing 50 μg/ml ampicillin and incubated overnight at 37°C. The seeded bacteria were treated with 1 mM isopropyl β-D-1-thiogalactopyranoside (Gold Biotechnology, St. Louis, MO, USA) and incubated at room temperature for approximately 24 h to induce dsRNAs. Age-synchronized wild-type N2 and daf-2(e1370) mutant animals were grown on RNAi plates from embryo to L4 stage at 20°C. Worms were then transferred onto RNAi plates containing 5 μM 5-fluoro-2’deoxyuridine (FUdR; Sigma, St. Louis, MO, USA), which prevents eggs from hatching, at young (day 1) adult stage, and transferred again to new FUdR-containing RNAi plates after 1 or 2 days. All lifespan assays were performed at 20°C as duplicates by two independent researchers. The survival of worms was determined by gently touching worms with a platinum wire. Worms that did not respond were counted as dead worms and removed from the plates. Worms that crawled off the plates, burrowed into the agar media, or displayed internal hatching or vulval rupture were censored, but included in the statistical analysis. Lifespan data from two independent lifespan experiments within the experimental sets were pooled and used for statistical analysis (see Dataset 1–Dataset 3). OASIS 2 (Online Application for Survival Analysis 2; https://sbi.postech.ac.kr/oasis2)22 was used for the statistical analysis of lifespan results. P values were calculated using long-rank (Mantel-Cox method) test.
Results
Knocking down each of several C. elegans α-arrestins marginally influenced lifespan
We measured the lifespan of wild-type and long-lived daf-2 mutant animals upon knocking down each of 24 of 29 genes encoding putative α-arrestin proteins (Figure 1A). We used daf-16 RNAi that largely suppressed the longevity of daf-2(-) mutants as a positive control (Figure 1B, Figure S1A, Table S1)17,18. We found that knockdown of each of individual α-arrestin genes tended to slightly reduce lifespan in wild-type or in daf-2(-) mutants (Figure 1B, Figure S1B–U, Table S1). Out of the 24 RNAi clones, RNAi targeting arrd (arrestin domain protein)-13, arrd-16, arrd-23, arrd-24 or arrd-25 in wild-type, and RNAi against arrd-1, arrd-2, arrd-5, arrd-24 or arrd-28 in daf-2(-) mutant animals decreased lifespan by more than 5% (Figure 1B–E, Figures S1B, D, K, Q, R, T, Table S1). Specifically, arrd-16 RNAi decreased lifespan in wild-type by 9%, and arrd-1 RNAi decreased lifespan in daf-2(-) mutants by 7% (Figure 1B and C). In addition, RNAi targeting arrd-24 decreased the lifespan of wild-type and daf-2 mutant animals by 11% and 6%, respectively (Figure 1E). In contrast, arrd-3 RNAi increased the lifespan of wild-type animals by 10% (Figure 1F). Overall, genetic inhibition of individual α-arrestin genes using RNAi appears to have minor effects on C. elegans lifespan.
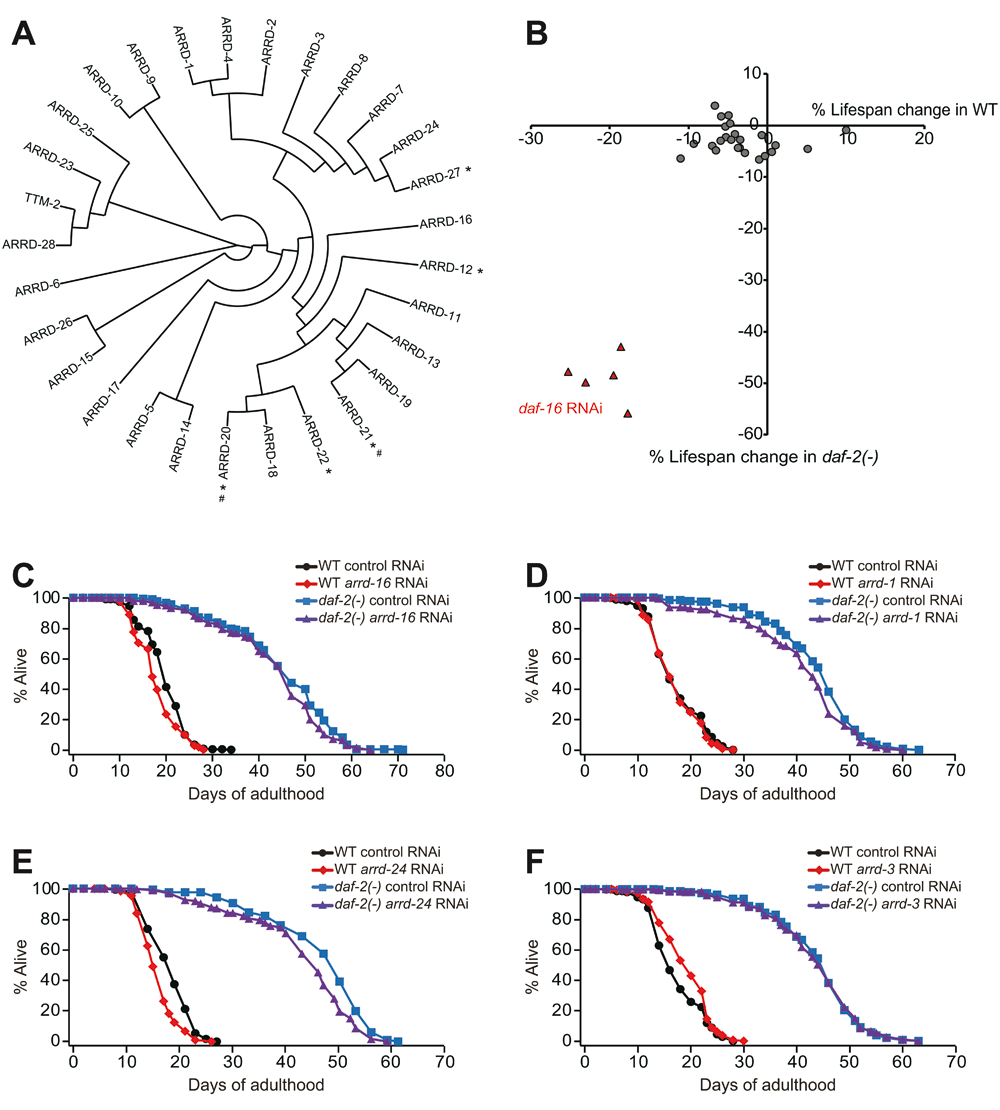
Figure 1. RNAi targeting each of Caenorhabditis elegans α-arrestins had small or no effects on lifespan.
(A) A phylogenetic tree showing C. elegans α-arrestin family members. Asterisks (*) indicate α-arrestins that were not examined in this study and number signs (#) indicate predicted pseudogenes. (B) Each circle represents percent mean lifespan change by treatment with individual α-arrestin gene RNAi in wild-type (WT) and daf-2(e1370) (daf-2(-)) mutants. Red triangles indicate lifespan changes by daf-16 RNAi, which was used as a positive control. (C–F) Lifespan curves of WT and daf-2(-) animals treated with RNAi targeting arrd-16 (C), arrd-1 (D), arrd- 24 (E), and arrd-3 (F). See Table S1 for statistical analysis.
| Set | Strain | Treatment | Days | At Risk | Dead | Censored | Percent Mortality | S_hat | Var S_hat | SE(S) | Survival Time | Percent Alive |
|---|
| 1 | N2 | Control RNAi | 0 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | Control RNAi | 1 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | Control RNAi | 2 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | Control RNAi | 4 | 240 | 1 | 15 | 0.42 | 0.996 | 0 | 0.004 | 0.017 | 99.6 |
| 1 | N2 | Control RNAi | 5 | 224 | 0 | 2 | 0.42 | 0.996 | 0 | 0.004 | 0.017 | 99.6 |
| 1 | N2 | Control RNAi | 6 | 222 | 2 | 19 | 1.31 | 0.987 | 0 | 0.008 | 0.07 | 98.7 |
| 1 | N2 | Control RNAi | 7 | 201 | 0 | 1 | 1.31 | 0.987 | 0 | 0.008 | 0.07 | 98.7 |
| 1 | N2 | Control RNAi | 8 | 200 | 2 | 10 | 2.3 | 0.977 | 0 | 0.01 | 0.149 | 97.7 |
| 1 | N2 | Control RNAi | 10 | 188 | 6 | 2 | 5.42 | 0.946 | 0 | 0.016 | 0.461 | 94.6 |
| 1 | N2 | Control RNAi | 11 | 180 | 3 | 10 | 7 | 0.93 | 0 | 0.018 | 0.635 | 93 |
| 1 | N2 | Control RNAi | 12 | 167 | 10 | 1 | 12.56 | 0.874 | 0.001 | 0.024 | 1.303 | 87.4 |
| 1 | N2 | Control RNAi | 14 | 156 | 44 | 4 | 37.23 | 0.628 | 0.001 | 0.036 | 4.756 | 62.8 |
| 1 | N2 | Control RNAi | 16 | 108 | 28 | 2 | 53.5 | 0.465 | 0.001 | 0.038 | 7.36 | 46.5 |
| 1 | N2 | Control RNAi | 18 | 78 | 21 | 1 | 66.02 | 0.34 | 0.001 | 0.036 | 9.613 | 34 |
| 1 | N2 | Control RNAi | 20 | 56 | 14 | 1 | 74.51 | 0.255 | 0.001 | 0.033 | 11.312 | 25.5 |
| 1 | N2 | Control RNAi | 22 | 41 | 5 | 0 | 77.62 | 0.224 | 0.001 | 0.032 | 11.996 | 22.4 |
| 1 | N2 | Control RNAi | 23 | 36 | 17 | 0 | 88.19 | 0.118 | 0.001 | 0.025 | 14.426 | 11.8 |
| 1 | N2 | Control RNAi | 24 | 19 | 5 | 0 | 91.3 | 0.087 | 0 | 0.022 | 15.172 | 8.7 |
| 1 | N2 | Control RNAi | 25 | 14 | 7 | 0 | 95.65 | 0.044 | 0 | 0.016 | 16.26 | 4.4 |
| 1 | N2 | Control RNAi | 26 | 7 | 3 | 0 | 97.51 | 0.025 | 0 | 0.012 | 16.745 | 2.5 |
| 1 | N2 | Control RNAi | 28 | 4 | 4 | 0 | 100 | 0 | 0 | 0 | 17.441 | 0 |
| 1 | N2 | daf-16 RNAi | 0 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | daf-16 RNAi | 1 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | daf-16 RNAi | 2 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | daf-16 RNAi | 4 | 240 | 2 | 28 | 0.83 | 0.992 | 0 | 0.006 | 0.033 | 99.2 |
| 1 | N2 | daf-16 RNAi | 5 | 210 | 0 | 2 | 0.83 | 0.992 | 0 | 0.006 | 0.033 | 99.2 |
| 1 | N2 | daf-16 RNAi | 6 | 208 | 1 | 20 | 1.31 | 0.987 | 0 | 0.008 | 0.062 | 98.7 |
| 1 | N2 | daf-16 RNAi | 7 | 187 | 0 | 5 | 1.31 | 0.987 | 0 | 0.008 | 0.062 | 98.7 |
| 1 | N2 | daf-16 RNAi | 8 | 182 | 4 | 7 | 3.48 | 0.965 | 0 | 0.013 | 0.235 | 96.5 |
| 1 | N2 | daf-16 RNAi | 10 | 171 | 9 | 1 | 8.56 | 0.914 | 0 | 0.021 | 0.743 | 91.4 |
| 1 | N2 | daf-16 RNAi | 11 | 161 | 37 | 9 | 29.57 | 0.704 | 0.001 | 0.034 | 3.055 | 70.4 |
| 1 | N2 | daf-16 RNAi | 12 | 115 | 10 | 2 | 35.7 | 0.643 | 0.001 | 0.036 | 3.79 | 64.3 |
| 1 | N2 | daf-16 RNAi | 14 | 103 | 48 | 5 | 65.66 | 0.343 | 0.001 | 0.037 | 7.985 | 34.3 |
| 1 | N2 | daf-16 RNAi | 16 | 50 | 25 | 3 | 82.83 | 0.172 | 0.001 | 0.031 | 10.732 | 17.2 |
| 1 | N2 | daf-16 RNAi | 18 | 22 | 12 | 0 | 92.2 | 0.078 | 0.001 | 0.023 | 12.418 | 7.8 |
| 1 | N2 | daf-16 RNAi | 20 | 10 | 7 | 0 | 97.66 | 0.023 | 0 | 0.013 | 13.51 | 2.3 |
| 1 | N2 | daf-16 RNAi | 22 | 3 | 3 | 0 | 100 | 0 | 0 | 0 | 14.025 | 0 |
| 1 | N2 | arrd-1 RNAi | 0 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-1 RNAi | 1 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-1 RNAi | 2 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-1 RNAi | 4 | 240 | 0 | 12 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-1 RNAi | 5 | 228 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-1 RNAi | 6 | 225 | 1 | 38 | 0.44 | 0.996 | 0 | 0.004 | 0.027 | 99.6 |
| 1 | N2 | arrd-1 RNAi | 7 | 186 | 0 | 1 | 0.44 | 0.996 | 0 | 0.004 | 0.027 | 99.6 |
| 1 | N2 | arrd-1 RNAi | 8 | 185 | 0 | 12 | 0.44 | 0.996 | 0 | 0.004 | 0.027 | 99.6 |
| 1 | N2 | arrd-1 RNAi | 10 | 173 | 2 | 0 | 1.6 | 0.984 | 0 | 0.009 | 0.142 | 98.4 |
| 1 | N2 | arrd-1 RNAi | 11 | 171 | 17 | 7 | 11.38 | 0.886 | 0.001 | 0.024 | 1.218 | 88.6 |
| 1 | N2 | arrd-1 RNAi | 12 | 147 | 5 | 2 | 14.39 | 0.856 | 0.001 | 0.027 | 1.58 | 85.6 |
| 1 | N2 | arrd-1 RNAi | 14 | 140 | 36 | 3 | 36.41 | 0.636 | 0.001 | 0.037 | 4.661 | 63.6 |
| 1 | N2 | arrd-1 RNAi | 16 | 101 | 25 | 1 | 52.15 | 0.479 | 0.002 | 0.039 | 7.18 | 47.9 |
| 1 | N2 | arrd-1 RNAi | 18 | 75 | 26 | 2 | 68.74 | 0.313 | 0.001 | 0.037 | 10.166 | 31.3 |
| 1 | N2 | arrd-1 RNAi | 20 | 47 | 10 | 1 | 75.39 | 0.246 | 0.001 | 0.034 | 11.496 | 24.6 |
| 1 | N2 | arrd-1 RNAi | 22 | 36 | 10 | 0 | 82.22 | 0.178 | 0.001 | 0.031 | 13.001 | 17.8 |
| 1 | N2 | arrd-1 RNAi | 23 | 26 | 14 | 0 | 91.8 | 0.082 | 0.001 | 0.022 | 15.202 | 8.2 |
| 1 | N2 | arrd-1 RNAi | 24 | 12 | 6 | 0 | 95.9 | 0.041 | 0 | 0.016 | 16.186 | 4.1 |
| 1 | N2 | arrd-1 RNAi | 25 | 6 | 1 | 0 | 96.58 | 0.034 | 0 | 0.015 | 16.357 | 3.4 |
| 1 | N2 | arrd-1 RNAi | 26 | 5 | 4 | 0 | 99.32 | 0.007 | 0 | 0.007 | 17.068 | 0.7 |
| 1 | N2 | arrd-1 RNAi | 28 | 1 | 1 | 0 | 100 | 0 | 0 | 0 | 17.26 | 0 |
| 1 | N2 | arrd-2 RNAi | 0 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-2 RNAi | 1 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-2 RNAi | 2 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-2 RNAi | 4 | 240 | 0 | 13 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-2 RNAi | 5 | 227 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-2 RNAi | 6 | 227 | 1 | 30 | 0.44 | 0.996 | 0 | 0.004 | 0.026 | 99.6 |
| 1 | N2 | arrd-2 RNAi | 7 | 196 | 0 | 0 | 0.44 | 0.996 | 0 | 0.004 | 0.026 | 99.6 |
| 1 | N2 | arrd-2 RNAi | 8 | 196 | 0 | 18 | 0.44 | 0.996 | 0 | 0.004 | 0.026 | 99.6 |
| 1 | N2 | arrd-2 RNAi | 10 | 178 | 3 | 3 | 2.12 | 0.979 | 0 | 0.011 | 0.194 | 97.9 |
| 1 | N2 | arrd-2 RNAi | 11 | 172 | 16 | 2 | 11.22 | 0.888 | 0.001 | 0.024 | 1.196 | 88.8 |
| 1 | N2 | arrd-2 RNAi | 12 | 154 | 4 | 0 | 13.53 | 0.865 | 0.001 | 0.026 | 1.473 | 86.5 |
| 1 | N2 | arrd-2 RNAi | 14 | 150 | 36 | 3 | 34.28 | 0.657 | 0.001 | 0.036 | 4.378 | 65.7 |
| 1 | N2 | arrd-2 RNAi | 16 | 111 | 22 | 0 | 47.31 | 0.527 | 0.001 | 0.038 | 6.462 | 52.7 |
| 1 | N2 | arrd-2 RNAi | 18 | 89 | 28 | 0 | 63.88 | 0.361 | 0.001 | 0.037 | 9.446 | 36.1 |
| 1 | N2 | arrd-2 RNAi | 20 | 61 | 20 | 0 | 75.73 | 0.243 | 0.001 | 0.033 | 11.814 | 24.3 |
| 1 | N2 | arrd-2 RNAi | 22 | 41 | 7 | 0 | 79.87 | 0.201 | 0.001 | 0.031 | 12.726 | 20.1 |
| 1 | N2 | arrd-2 RNAi | 23 | 34 | 21 | 0 | 92.3 | 0.077 | 0 | 0.02 | 15.585 | 7.7 |
| 1 | N2 | arrd-2 RNAi | 24 | 13 | 5 | 0 | 95.26 | 0.047 | 0 | 0.016 | 16.296 | 4.7 |
| 1 | N2 | arrd-2 RNAi | 25 | 8 | 2 | 0 | 96.45 | 0.036 | 0 | 0.014 | 16.592 | 3.6 |
| 1 | N2 | arrd-2 RNAi | 26 | 6 | 4 | 0 | 98.82 | 0.012 | 0 | 0.008 | 17.208 | 1.2 |
| 1 | N2 | arrd-2 RNAi | 28 | 2 | 2 | 0 | 100 | 0 | 0 | 0 | 17.539 | 0 |
| 1 | N2 | arrd-3 RNAi | 0 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-3 RNAi | 1 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-3 RNAi | 2 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-3 RNAi | 4 | 240 | 0 | 6 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-3 RNAi | 5 | 234 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-3 RNAi | 6 | 233 | 2 | 23 | 0.86 | 0.991 | 0 | 0.006 | 0.052 | 99.1 |
| 1 | N2 | arrd-3 RNAi | 7 | 208 | 0 | 0 | 0.86 | 0.991 | 0 | 0.006 | 0.052 | 99.1 |
| 1 | N2 | arrd-3 RNAi | 8 | 208 | 2 | 13 | 1.81 | 0.982 | 0 | 0.009 | 0.128 | 98.2 |
| 1 | N2 | arrd-3 RNAi | 10 | 193 | 2 | 1 | 2.83 | 0.972 | 0 | 0.011 | 0.23 | 97.2 |
| 1 | N2 | arrd-3 RNAi | 11 | 190 | 7 | 0 | 6.41 | 0.936 | 0 | 0.017 | 0.623 | 93.6 |
| 1 | N2 | arrd-3 RNAi | 12 | 183 | 4 | 1 | 8.45 | 0.915 | 0 | 0.02 | 0.869 | 91.5 |
| 1 | N2 | arrd-3 RNAi | 14 | 178 | 27 | 8 | 22.34 | 0.777 | 0.001 | 0.03 | 2.813 | 77.7 |
| 1 | N2 | arrd-3 RNAi | 16 | 143 | 20 | 3 | 33.2 | 0.668 | 0.001 | 0.034 | 4.551 | 66.8 |
| 1 | N2 | arrd-3 RNAi | 18 | 120 | 25 | 1 | 47.12 | 0.529 | 0.001 | 0.037 | 7.056 | 52.9 |
| 1 | N2 | arrd-3 RNAi | 20 | 94 | 18 | 0 | 57.24 | 0.428 | 0.001 | 0.037 | 9.081 | 42.8 |
| 1 | N2 | arrd-3 RNAi | 22 | 76 | 18 | 0 | 67.37 | 0.326 | 0.001 | 0.035 | 11.309 | 32.6 |
| 1 | N2 | arrd-3 RNAi | 23 | 58 | 32 | 1 | 85.37 | 0.146 | 0.001 | 0.026 | 15.449 | 14.6 |
| 1 | N2 | arrd-3 RNAi | 24 | 25 | 10 | 0 | 91.22 | 0.088 | 0 | 0.021 | 16.853 | 8.8 |
| 1 | N2 | arrd-3 RNAi | 25 | 15 | 5 | 0 | 94.15 | 0.059 | 0 | 0.018 | 17.585 | 5.9 |
| 1 | N2 | arrd-3 RNAi | 26 | 10 | 4 | 0 | 96.49 | 0.035 | 0 | 0.014 | 18.193 | 3.5 |
| 1 | N2 | arrd-3 RNAi | 28 | 6 | 5 | 0 | 99.41 | 0.006 | 0 | 0.006 | 19.012 | 0.6 |
| 1 | N2 | arrd-3 RNAi | 30 | 1 | 1 | 0 | 100 | 0 | 0 | 0 | 19.188 | 0 |
| 1 | N2 | arrd-4 RNAi | 0 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-4 RNAi | 1 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-4 RNAi | 2 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-4 RNAi | 4 | 240 | 0 | 15 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-4 RNAi | 5 | 225 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-4 RNAi | 6 | 223 | 1 | 22 | 0.45 | 0.996 | 0 | 0.004 | 0.027 | 99.6 |
| 1 | N2 | arrd-4 RNAi | 7 | 200 | 0 | 0 | 0.45 | 0.996 | 0 | 0.004 | 0.027 | 99.6 |
| 1 | N2 | arrd-4 RNAi | 8 | 200 | 2 | 15 | 1.44 | 0.986 | 0 | 0.008 | 0.107 | 98.6 |
| 1 | N2 | arrd-4 RNAi | 10 | 183 | 4 | 5 | 3.6 | 0.964 | 0 | 0.013 | 0.322 | 96.4 |
| 1 | N2 | arrd-4 RNAi | 11 | 174 | 20 | 3 | 14.68 | 0.853 | 0.001 | 0.026 | 1.541 | 85.3 |
| 1 | N2 | arrd-4 RNAi | 12 | 151 | 6 | 2 | 18.07 | 0.819 | 0.001 | 0.029 | 1.948 | 81.9 |
| 1 | N2 | arrd-4 RNAi | 14 | 143 | 24 | 4 | 31.82 | 0.682 | 0.001 | 0.035 | 3.873 | 68.2 |
| 1 | N2 | arrd-4 RNAi | 16 | 115 | 17 | 3 | 41.9 | 0.581 | 0.001 | 0.037 | 5.485 | 58.1 |
| 1 | N2 | arrd-4 RNAi | 18 | 95 | 17 | 1 | 52.3 | 0.477 | 0.001 | 0.038 | 7.357 | 47.7 |
| 1 | N2 | arrd-4 RNAi | 20 | 77 | 21 | 0 | 65.31 | 0.347 | 0.001 | 0.037 | 9.959 | 34.7 |
| 1 | N2 | arrd-4 RNAi | 22 | 56 | 11 | 0 | 72.12 | 0.279 | 0.001 | 0.035 | 11.458 | 27.9 |
| 1 | N2 | arrd-4 RNAi | 23 | 45 | 22 | 0 | 85.75 | 0.142 | 0.001 | 0.027 | 14.593 | 14.2 |
| 1 | N2 | arrd-4 RNAi | 24 | 23 | 5 | 0 | 88.85 | 0.112 | 0.001 | 0.025 | 15.336 | 11.2 |
| 1 | N2 | arrd-4 RNAi | 25 | 18 | 4 | 0 | 91.33 | 0.087 | 0 | 0.022 | 15.956 | 8.7 |
| 1 | N2 | arrd-4 RNAi | 26 | 14 | 5 | 0 | 94.42 | 0.056 | 0 | 0.018 | 16.761 | 5.6 |
| 1 | N2 | arrd-4 RNAi | 28 | 9 | 8 | 0 | 99.38 | 0.006 | 0 | 0.006 | 18.149 | 0.6 |
| 1 | N2 | arrd-4 RNAi | 30 | 1 | 1 | 0 | 100 | 0 | 0 | 0 | 18.335 | 0 |
| 1 | N2 | arrd-5 RNAi | 0 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-5 RNAi | 1 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-5 RNAi | 2 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-5 RNAi | 4 | 240 | 0 | 9 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-5 RNAi | 5 | 231 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | N2 | arrd-5 RNAi | 6 | 228 | 5 | 27 | 2.19 | 0.978 | 0 | 0.01 | 0.132 | 97.8 |
| 1 | N2 | arrd-5 RNAi | 7 | 196 | 1 | 1 | 2.69 | 0.973 | 0 | 0.011 | 0.167 | 97.3 |
| 1 | N2 | arrd-5 RNAi | 8 | 194 | 0 | 15 | 2.69 | 0.973 | 0 | 0.011 | 0.167 | 97.3 |
| 1 | N2 | arrd-5 RNAi | 10 | 179 | 3 | 3 | 4.32 | 0.957 | 0 | 0.014 | 0.33 | 95.7 |
| 1 | N2 | arrd-5 RNAi | 11 | 173 | 9 | 7 | 9.3 | 0.907 | 0 | 0.021 | 0.877 | 90.7 |
| 1 | N2 | arrd-5 RNAi | 12 | 157 | 2 | 0 | 10.46 | 0.895 | 0 | 0.022 | 1.016 | 89.5 |
| 1 | N2 | arrd-5 RNAi | 14 | 155 | 41 | 6 | 34.14 | 0.659 | 0.001 | 0.036 | 4.332 | 65.9 |
| 1 | N2 | arrd-5 RNAi | 16 | 108 | 19 | 2 | 45.73 | 0.543 | 0.001 | 0.038 | 6.186 | 54.3 |
| 1 | N2 | arrd-5 RNAi | 18 | 87 | 25 | 1 | 61.32 | 0.387 | 0.001 | 0.038 | 8.993 | 38.7 |
| 1 | N2 | arrd-5 RNAi | 20 | 61 | 30 | 2 | 80.34 | 0.197 | 0.001 | 0.031 | 12.797 | 19.7 |
| 1 | N2 | arrd-5 RNAi | 22 | 29 | 9 | 0 | 86.44 | 0.136 | 0.001 | 0.027 | 14.139 | 13.6 |
| 1 | N2 | arrd-5 RNAi | 23 | 20 | 11 | 1 | 93.9 | 0.061 | 0 | 0.019 | 15.854 | 6.1 |
| 1 | N2 | arrd-5 RNAi | 24 | 8 | 2 | 0 | 95.43 | 0.046 | 0 | 0.017 | 16.22 | 4.6 |
| 1 | N2 | arrd-5 RNAi | 25 | 6 | 4 | 0 | 98.48 | 0.015 | 0 | 0.011 | 16.982 | 1.5 |
| 1 | N2 | arrd-5 RNAi | 26 | 2 | 1 | 0 | 99.24 | 0.008 | 0 | 0.008 | 17.181 | 0.8 |
| 1 | N2 | arrd-5 RNAi | 28 | 1 | 1 | 0 | 100 | 0 | 0 | 0 | 17.394 | 0 |
| 1 | daf-2(e1370) | Control RNAi | 0 | 210 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | Control RNAi | 1 | 210 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | Control RNAi | 2 | 210 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | Control RNAi | 4 | 210 | 0 | 6 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | Control RNAi | 7 | 204 | 0 | 8 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | Control RNAi | 8 | 196 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | Control RNAi | 10 | 196 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | Control RNAi | 11 | 192 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | Control RNAi | 13 | 189 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | Control RNAi | 14 | 185 | 1 | 3 | 0.54 | 0.995 | 0 | 0.005 | 0.076 | 99.5 |
| 1 | daf-2(e1370) | Control RNAi | 16 | 181 | 2 | 1 | 1.64 | 0.984 | 0 | 0.009 | 0.252 | 98.4 |
| 1 | daf-2(e1370) | Control RNAi | 18 | 178 | 0 | 0 | 1.64 | 0.984 | 0 | 0.009 | 0.252 | 98.4 |
| 1 | daf-2(e1370) | Control RNAi | 19 | 178 | 1 | 0 | 2.19 | 0.978 | 0 | 0.011 | 0.357 | 97.8 |
| 1 | daf-2(e1370) | Control RNAi | 20 | 177 | 0 | 6 | 2.19 | 0.978 | 0 | 0.011 | 0.357 | 97.8 |
| 1 | daf-2(e1370) | Control RNAi | 22 | 171 | 1 | 0 | 2.76 | 0.972 | 0 | 0.012 | 0.482 | 97.2 |
| 1 | daf-2(e1370) | Control RNAi | 23 | 170 | 0 | 2 | 2.76 | 0.972 | 0 | 0.012 | 0.482 | 97.2 |
| 1 | daf-2(e1370) | Control RNAi | 25 | 168 | 2 | 17 | 3.92 | 0.961 | 0 | 0.015 | 0.772 | 96.1 |
| 1 | daf-2(e1370) | Control RNAi | 28 | 149 | 4 | 2 | 6.5 | 0.935 | 0 | 0.019 | 1.494 | 93.5 |
| 1 | daf-2(e1370) | Control RNAi | 30 | 143 | 0 | 2 | 6.5 | 0.935 | 0 | 0.019 | 1.494 | 93.5 |
| 1 | daf-2(e1370) | Control RNAi | 31 | 141 | 7 | 0 | 11.14 | 0.889 | 0.001 | 0.025 | 2.933 | 88.9 |
| 1 | daf-2(e1370) | Control RNAi | 33 | 134 | 1 | 0 | 11.81 | 0.882 | 0.001 | 0.026 | 3.152 | 88.2 |
| 1 | daf-2(e1370) | Control RNAi | 34 | 133 | 6 | 1 | 15.78 | 0.842 | 0.001 | 0.029 | 4.504 | 84.2 |
| 1 | daf-2(e1370) | Control RNAi | 36 | 126 | 2 | 1 | 17.12 | 0.829 | 0.001 | 0.03 | 4.986 | 82.9 |
| 1 | daf-2(e1370) | Control RNAi | 37 | 123 | 7 | 1 | 21.84 | 0.782 | 0.001 | 0.033 | 6.731 | 78.2 |
| 1 | daf-2(e1370) | Control RNAi | 38 | 115 | 4 | 1 | 24.56 | 0.754 | 0.001 | 0.035 | 7.764 | 75.4 |
| 1 | daf-2(e1370) | Control RNAi | 40 | 110 | 10 | 0 | 31.42 | 0.686 | 0.001 | 0.038 | 10.507 | 68.6 |
| 1 | daf-2(e1370) | Control RNAi | 41 | 100 | 3 | 0 | 33.47 | 0.665 | 0.001 | 0.038 | 11.351 | 66.5 |
| 1 | daf-2(e1370) | Control RNAi | 43 | 97 | 12 | 0 | 41.7 | 0.583 | 0.002 | 0.04 | 14.89 | 58.3 |
| 1 | daf-2(e1370) | Control RNAi | 44 | 85 | 6 | 1 | 45.82 | 0.542 | 0.002 | 0.041 | 16.701 | 54.2 |
| 1 | daf-2(e1370) | Control RNAi | 46 | 78 | 23 | 0 | 61.79 | 0.382 | 0.002 | 0.04 | 24.05 | 38.2 |
| 1 | daf-2(e1370) | Control RNAi | 49 | 55 | 26 | 0 | 79.86 | 0.201 | 0.001 | 0.033 | 32.9 | 20.1 |
| 1 | daf-2(e1370) | Control RNAi | 51 | 29 | 10 | 0 | 86.8 | 0.132 | 0.001 | 0.028 | 36.442 | 13.2 |
| 1 | daf-2(e1370) | Control RNAi | 52 | 19 | 6 | 0 | 90.97 | 0.09 | 0.001 | 0.024 | 38.61 | 9 |
| 1 | daf-2(e1370) | Control RNAi | 54 | 13 | 5 | 0 | 94.44 | 0.056 | 0 | 0.019 | 40.485 | 5.6 |
| 1 | daf-2(e1370) | Control RNAi | 55 | 8 | 3 | 0 | 96.53 | 0.035 | 0 | 0.015 | 41.631 | 3.5 |
| 1 | daf-2(e1370) | Control RNAi | 57 | 5 | 2 | 0 | 97.92 | 0.021 | 0 | 0.012 | 42.423 | 2.1 |
| 1 | daf-2(e1370) | Control RNAi | 60 | 3 | 2 | 0 | 99.31 | 0.007 | 0 | 0.007 | 43.257 | 0.7 |
| 1 | daf-2(e1370) | Control RNAi | 63 | 1 | 1 | 0 | 100 | 0 | 0 | 0 | 43.694 | 0 |
| 1 | daf-2(e1370) | daf-16 RNAi | 0 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | daf-16 RNAi | 1 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | daf-16 RNAi | 2 | 240 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | daf-16 RNAi | 4 | 240 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 100 |
| 1 | daf-2(e1370) | daf-16 RNAi | 7 | 237 | 4 | 12 | 1.69 | 0.983 | 0 | 0.008 | 0.118 | 98.3 |
| 1 | daf-2(e1370) | daf-16 RNAi | 8 | 221 | 0 | 1 | 1.69 | 0.983 | 0 | 0.008 | 0.118 | 98.3 |
| 1 | daf-2(e1370) | daf-16 RNAi | 10 | 220 | 4 | 2 | 3.48 | 0.965 | 0 | 0.012 | 0.297 | 96.5 |
| 1 | daf-2(e1370) | daf-16 RNAi | 11 | 214 | 4 | 1 | 5.28 | 0.947 | 0 | 0.015 | 0.495 | 94.7 |
| 1 | daf-2(e1370) | daf-16 RNAi | 13 | 209 | 2 | 2 | 6.19 | 0.938 | 0 | 0.016 | 0.613 | 93.8 |
| 1 | daf-2(e1370) | daf-16 RNAi | 14 | 205 | 8 | 2 | 9.85 | 0.902 | 0 | 0.02 | 1.126 | 90.2 |
| 1 | daf-2(e1370) | daf-16 RNAi | 16 | 195 | 11 | 1 | 14.93 | 0.851 | 0.001 | 0.024 | 1.939 | 85.1 |
| 1 | daf-2(e1370) | daf-16 RNAi | 18 | 183 | 10 | 0 | 19.58 | 0.804 | 0.001 | 0.027 | 2.776 | 80.4 |
| 1 | daf-2(e1370) | daf-16 RNAi | 19 | 173 | 16 | 0 | 27.02 | 0.73 | 0.001 | 0.03 | 4.189 | 73 |
| 1 | daf-2(e1370) | daf-16 RNAi | 20 | 157 | 18 | 3 | 35.39 | 0.646 | 0.001 | 0.032 | 5.863 | 64.6 |
| 1 | daf-2(e1370) | daf-16 RNAi | 22 | 136 | 17 | 1 | 43.46 | 0.565 | 0.001 | 0.034 | 7.64 | 56.5 |
| 1 | daf-2(e1370) | daf-16 RNAi | 23 | 118 | 33 | 0 | 59.27 | 0.407 | 0.001 | 0.034 | 11.276 | 40.7 |
| 1 | daf-2(e1370) | daf-16 RNAi | 25 | 85 | 30 | 0 | 73.65 | 0.264 | 0.001 | 0.03 | 14.87 | 26.4 |
| This is a portion of the data; to view all the data, please download the file. |
Dataset 1.Kaplan-Meier estimator of RNAi lifespan experiments.
Kaplan-Meier estimate values were calculated from pooled lifespan data of two independent lifespan experiments using OASIS2 (https://sbi.postech.ac.kr/oasis2/). ‘At risk’ indicates the number of individuals at risk just prior to a specific time point. ‘S_hat’ indicates Kaplan-Meier estimate of survival function, ‘Var S_hat’ indicates variance of ‘S_hat’, and ‘SE(S)’ indicates standard error of ‘S_hat’.| Set | Strain | Treatment | Number of subjects | Mean survival days | Standard error of mean survival | 95 percent confidence interval of mean | Age in days at 25 percent mortailty | Age in days at 50 percent mortailty | Age in days at 75 percent mortailty | Age in days at 90 percent mortailty | Age in days at 100 percent mortailty | 95 percent median confidence interval | 25 percent linear interpolation of mortality curve | 50 percent linear interpolation of mortality curve | 75 percent linear interpolation of mortality curve | 90 percent linear interpolation of mortality curve | 100 percent linear interpolation of mortality curve |
|---|
| 1 | N2 | control RNAi | 240 | 17.44 | 0.35 | 16.75 ~ 18.13 | 14 | 16 | 22 | 24 | 28 | 16.0 ~ 16.0 | 13.01 | 15.57 | 20.31 | 23.58 | 28 |
| 1 | N2 | daf-16 RNAi | 240 | 14.03 | 0.25 | 13.54 ~ 14.51 | 11 | 14 | 16 | 18 | 22 | - ~ - | 10.78 | 12.95 | 15.09 | 17.53 | 22 |
| 1 | N2 | arrd-1 RNAi | 240 | 17.26 | 0.34 | 16.60 ~ 17.92 | 14 | 16 | 20 | 23 | 28 | 16.0 ~ 16.0 | 12.96 | 15.73 | 19.88 | 22.81 | 28 |
| 1 | N2 | arrd-2 RNAi | 240 | 17.54 | 0.33 | 16.89 ~ 18.19 | 14 | 18 | 20 | 23 | 28 | 16.0 ~ 16.0 | 13.11 | 16.32 | 19.88 | 22.81 | 28 |
| 1 | N2 | arrd-3 RNAi | 240 | 19.19 | 0.34 | 18.53 ~ 19.85 | 16 | 20 | 23 | 24 | 30 | 18.0 ~ 18.0 | 14.49 | 18.57 | 22.42 | 23.79 | 30 |
| 1 | N2 | arrd-4 RNAi | 240 | 18.34 | 0.39 | 17.56 ~ 19.11 | 14 | 18 | 23 | 25 | 30 | 18.0 ~ 18.0 | 13.01 | 17.56 | 22.21 | 24.46 | 30 |
| 1 | N2 | arrd-5 RNAi | 240 | 17.39 | 0.32 | 16.76 ~ 18.02 | 14 | 18 | 20 | 23 | 28 | 16.0 ~ 16.0 | 13.23 | 16.55 | 19.44 | 22.48 | 28 |
| 1 | daf-2(e1370) | control RNAi | 210 | 43.69 | 0.69 | 42.35 ~ 45.04 | 40 | 46 | 49 | 52 | 63 | 44.0 ~ 44.0 | 38.13 | 44.52 | 48.19 | 51.77 | 63 |
| 1 | daf-2(e1370) | daf-16 RNAi | 240 | 22.52 | 0.37 | 21.79 ~ 23.25 | 19 | 23 | 28 | 28 | 34 | 22.0 ~ 22.0 | 18.73 | 22.41 | 25.22 | 27.63 | 34 |
| 1 | daf-2(e1370) | arrd-1 RNAi | 180 | 40.81 | 0.85 | 39.15 ~ 42.47 | 36 | 43 | 46 | 52 | 60 | 41.0 ~ 44.0 | 34.76 | 42.49 | 45.88 | 51.34 | 60 |
| 1 | daf-2(e1370) | arrd-2 RNAi | 210 | 41.47 | 0.66 | 40.18 ~ 42.76 | 36 | 44 | 46 | 51 | 57 | 43.0 ~ 43.0 | 35.75 | 43.17 | 45.9 | 50.15 | 57 |
| 1 | daf-2(e1370) | arrd-3 RNAi | 240 | 43.32 | 0.65 | 42.04 ~ 44.59 | 38 | 46 | 49 | 52 | 63 | 43.0 ~ 44.0 | 37.42 | 44.03 | 48.52 | 51.77 | 63 |
| 1 | daf-2(e1370) | arrd-4 RNAi | 240 | 41.74 | 0.65 | 40.45 ~ 43.02 | 37 | 43 | 49 | 52 | 60 | 41.0 ~ 43.0 | 36.34 | 42.36 | 47.09 | 51.55 | 60 |
| 1 | daf-2(e1370) | arrd-5 RNAi | 240 | 41.11 | 0.6 | 39.93 ~ 42.28 | 37 | 43 | 46 | 51 | 57 | 41.0 ~ 43.0 | 36.08 | 41.49 | 45.81 | 50.26 | 57 |
| 2 | N2 | control RNAi | 240 | 21.66 | 0.35 | 20.97 ~ 22.36 | 18 | 22 | 24 | 27 | 34 | 21.0 ~ 21.0 | 17.01 | 21 | 23.94 | 26.51 | 34 |
| 2 | N2 | daf-16 RNAi | 240 | 17.63 | 0.31 | 17.02 ~ 18.24 | 15 | 18 | 22 | 24 | 27 | 18.0 ~ 18.0 | 12.51 | 16.89 | 21.09 | 22.51 | 27 |
| 2 | N2 | arrd-6 RNAi | 240 | 20.51 | 0.43 | 19.68 ~ 21.35 | 15 | 19 | 24 | 30 | 37 | 19.0 ~ 21.0 | 14.27 | 18.98 | 23.93 | 27.15 | 37 |
| 2 | N2 | arrd-7 RNAi | 240 | 20.92 | 0.39 | 20.17 ~ 21.68 | 15 | 21 | 24 | 27 | 34 | 19.0 ~ 21.0 | 14.77 | 19.8 | 23.81 | 26.85 | 34 |
| 2 | N2 | arrd-8 RNAi | 240 | 20.52 | 0.42 | 19.69 ~ 21.35 | 15 | 21 | 24 | 30 | 30 | 19.0 ~ 21.0 | 13.94 | 19.31 | 23.77 | 27.72 | 30 |
| 2 | N2 | arrd-9 RNAi | 240 | 20.77 | 0.39 | 20.00 ~ 21.54 | 15 | 21 | 24 | 27 | 34 | 19.0 ~ 19.0 | 14.71 | 19.27 | 23.85 | 26.91 | 34 |
| 2 | N2 | arrd-10 RNAi | 240 | 20.22 | 0.41 | 19.42 ~ 21.02 | 15 | 21 | 24 | 27 | 30 | 19.0 ~ 21.0 | 13.79 | 19.6 | 23.46 | 26.53 | 30 |
| 2 | daf-2(e1370) | control RNAi | 240 | 44.31 | 0.73 | 42.88 ~ 45.75 | 39 | 48 | 51 | 57 | 62 | 44.0 ~ 45.0 | 37.78 | 45.29 | 50.37 | 55.18 | 62 |
| 2 | daf-2(e1370) | daf-16 RNAi | 241 | 25.28 | 0.41 | 24.47 ~ 26.09 | 21 | 24 | 30 | 34 | 40 | 24.0 ~ 24.0 | 20.6 | 23.58 | 27.85 | 33.09 | 40 |
| 2 | daf-2(e1370) | arrd-6 RNAi | 240 | 44.25 | 0.66 | 42.95 ~ 45.54 | 39 | 45 | 51 | 55 | 62 | 44.0 ~ 45.0 | 38.29 | 44.79 | 49.25 | 54.51 | 62 |
| 2 | daf-2(e1370) | arrd-7 RNAi | 240 | 43.09 | 0.74 | 41.64 ~ 44.55 | 37 | 45 | 51 | 55 | 62 | 44.0 ~ 45.0 | 36.15 | 44.52 | 49.54 | 54.84 | 62 |
| 2 | daf-2(e1370) | arrd-8 RNAi | 240 | 43.33 | 0.66 | 42.04 ~ 44.61 | 39 | 45 | 48 | 54 | 62 | 44.0 ~ 45.0 | 38.9 | 44.06 | 47.99 | 52.18 | 62 |
| 2 | daf-2(e1370) | arrd-9 RNAi | 240 | 43.59 | 0.66 | 42.30 ~ 44.88 | 40 | 45 | 51 | 54 | 62 | 44.0 ~ 45.0 | 39.41 | 44.68 | 48.43 | 52.92 | 62 |
| 2 | daf-2(e1370) | arrd-10 RNAi | 240 | 46.02 | 0.75 | 44.56 ~ 47.48 | 40 | 48 | 55 | 59 | 66 | 45.0 ~ 45.0 | 39.45 | 46.23 | 54.18 | 57.44 | 66 |
| 3 | N2 | control RNAi | 240 | 19.83 | 0.32 | 19.21 ~ 20.45 | 17 | 20 | 24 | 24 | 34 | - ~ - | 16.35 | 19.25 | 22.42 | 23.98 | 34 |
| 3 | N2 | daf-16 RNAi | 240 | 14.82 | 0.22 | 14.40 ~ 15.25 | 12 | 14 | 17 | 18 | 22 | 14.0 ~ 14.0 | 11.53 | 13.92 | 16.71 | 17.98 | 22 |
| 3 | N2 | arrd-13 RNAi | 210 | 18.67 | 0.3 | 18.08 ~ 19.27 | 16 | 18 | 22 | 24 | 30 | 18.0 ~ 18.0 | 15.38 | 17.96 | 20.24 | 23.29 | 30 |
| 3 | N2 | arrd-14 RNAi | 240 | 19.13 | 0.3 | 18.53 ~ 19.72 | 16 | 20 | 22 | 24 | 28 | 18.0 ~ 18.0 | 15.93 | 18.58 | 21.53 | 23.73 | 28 |
| 3 | N2 | arrd-15 RNAi | 240 | 18.86 | 0.32 | 18.23 ~ 19.49 | 14 | 20 | 22 | 26 | 30 | 18.0 ~ 18.0 | 13.76 | 18.24 | 20.83 | 24.95 | 30 |
| 3 | N2 | arrd-16 RNAi | 240 | 17.98 | 0.31 | 17.38 ~ 18.58 | 14 | 17 | 20 | 24 | 28 | 17.0 ~ 17.0 | 13.36 | 16.89 | 19.81 | 23.92 | 28 |
| 3 | N2 | arrd-18 RNAi | 240 | 18.89 | 0.33 | 18.25 ~ 19.54 | 14 | 18 | 22 | 26 | 32 | 17.0 ~ 18.0 | 13.89 | 17.94 | 21.14 | 25.05 | 32 |
| 3 | daf-2(e1370) | control RNAi | 210 | 44.89 | 0.85 | 43.23 ~ 46.55 | 38 | 47 | 53 | 58 | 71 | 44.0 ~ 47.0 | 37.86 | 45.39 | 52.85 | 57.13 | 71 |
| 3 | daf-2(e1370) | daf-16 RNAi | 240 | 23.45 | 0.33 | 22.80 ~ 24.11 | 20 | 24 | 26 | 28 | 38 | 24.0 ~ 24.0 | 18.94 | 23.2 | 25.67 | 27.83 | 38 |
| 3 | daf-2(e1370) | arrd-13 RNAi | 210 | 45.69 | 0.9 | 43.92 ~ 47.46 | 40 | 47 | 53 | 59 | 70 | 44.0 ~ 47.0 | 38.05 | 46.07 | 52.35 | 57.43 | 70 |
| 3 | daf-2(e1370) | arrd-14 RNAi | 240 | 42.94 | 0.84 | 41.30 ~ 44.58 | 37 | 44 | 50 | 56 | 67 | 44.0 ~ 44.0 | 36.09 | 43.51 | 49.94 | 54.3 | 67 |
| 3 | daf-2(e1370) | arrd-15 RNAi | 240 | 45.79 | 0.67 | 44.47 ~ 47.11 | 41 | 47 | 51 | 56 | 67 | - ~ - | 40.15 | 45.44 | 50.73 | 55.53 | 67 |
| 3 | daf-2(e1370) | arrd-16 RNAi | 240 | 43.29 | 0.82 | 41.68 ~ 44.89 | 37 | 47 | 51 | 56 | 64 | 44.0 ~ 44.0 | 36.65 | 44.82 | 50.49 | 54.23 | 64 |
| 3 | daf-2(e1370) | arrd-18 RNAi | 180 | 45.1 | 0.95 | 43.24 ~ 46.96 | 38 | 47 | 53 | 59 | 64 | 44.0 ~ 47.0 | 37.63 | 45.44 | 52.26 | 58.54 | 64 |
| 4 | N2 | control RNAi | 240 | 18.04 | 0.28 | 17.49 ~ 18.59 | 15 | 18 | 21 | 23 | 29 | 17.0 ~ 18.0 | 14.13 | 17.43 | 20.28 | 22.45 | 29 |
| 4 | N2 | daf-16 RNAi | 240 | 14.85 | 0.21 | 14.44 ~ 15.26 | 12 | 15 | 17 | 18 | 23 | 14.0 ~ 14.0 | 11.73 | 14.24 | 16.38 | 17.98 | 23 |
| 4 | N2 | arrd-23 RNAi | 240 | 16.78 | 0.3 | 16.19 ~ 17.38 | 14 | 17 | 19 | 23 | 27 | 15.0 ~ 17.0 | 13.03 | 16.15 | 18.99 | 21.81 | 27 |
| 4 | N2 | arrd-24 RNAi | 240 | 16.05 | 0.22 | 15.61 ~ 16.49 | 14 | 15 | 18 | 21 | 26 | 15.0 ~ 15.0 | 12.82 | 14.91 | 17.16 | 19.85 | 26 |
| 4 | N2 | arrd-25 RNAi | 240 | 16.86 | 0.27 | 16.32 ~ 17.39 | 15 | 17 | 19 | 21 | 26 | - ~ - | 12.94 | 15.94 | 18.56 | 20.74 | 26 |
| 4 | N2 | arrd-26 RNAi | 240 | 17.22 | 0.28 | 16.66 ~ 17.78 | 15 | 17 | 19 | 23 | 29 | 15.0 ~ 15.0 | 12.86 | 15.37 | 18.84 | 22.7 | 29 |
| 4 | N2 | arrd-28 RNAi | 240 | 17.53 | 0.27 | 17.00 ~ 18.07 | 15 | 17 | 21 | 23 | 29 | 17.0 ~ 17.0 | 13.4 | 16.79 | 19.31 | 21.92 | 29 |
| 4 | daf-2(e1370) | control RNAi | 210 | 45.93 | 0.75 | 44.45 ~ 47.40 | 39 | 50 | 53 | 56 | 61 | 49.0 ~ 49.0 | 38.88 | 49.03 | 52.68 | 55.15 | 61 |
| 4 | daf-2(e1370) | daf-16 RNAi | 240 | 20.29 | 0.35 | 19.60 ~ 20.97 | 18 | 21 | 24 | 27 | 29 | 18.0 ~ 18.0 | 15.32 | 18.75 | 23.43 | 26.7 | 29 |
| 4 | daf-2(e1370) | arrd-23 RNAi | 240 | 44.15 | 0.7 | 42.78 ~ 45.51 | 37 | 47 | 52 | 56 | 60 | 46.0 ~ 47.0 | 36.8 | 46.71 | 51.15 | 53.44 | 60 |
| 4 | daf-2(e1370) | arrd-24 RNAi | 211 | 43 | 0.75 | 41.52 ~ 44.47 | 39 | 46 | 50 | 53 | 59 | 46.0 ~ 46.0 | 38.29 | 44.82 | 49.36 | 52.81 | 59 |
| 4 | daf-2(e1370) | arrd-25 RNAi | 240 | 43.74 | 0.64 | 42.47 ~ 45.00 | 40 | 46 | 50 | 53 | 59 | 46.0 ~ 46.0 | 39.53 | 45.97 | 49.47 | 52.41 | 59 |
| 4 | daf-2(e1370) | arrd-26 RNAi | 241 | 44.67 | 0.64 | 43.42 ~ 45.93 | 39 | 47 | 52 | 53 | 59 | 46.0 ~ 47.0 | 38.39 | 46.61 | 50.32 | 52.94 | 59 |
| 4 | daf-2(e1370) | arrd-28 RNAi | 150 | 43.52 | 0.85 | 41.85 ~ 45.19 | 37 | 47 | 50 | 53 | 56 | 46.0 ~ 47.0 | 36.86 | 46.13 | 49.65 | 52.53 | 56 |
| 5 | N2 | control RNAi | 240 | 20.31 | 0.43 | 19.47 ~ 21.16 | 15 | 21 | 24 | 28 | 38 | 19.0 ~ 19.0 | 15 | 19.76 | 23.86 | 27.2 | 38 |
| 5 | N2 | daf-16 RNAi | 240 | 15.63 | 0.35 | 14.95 ~ 16.31 | 12 | 15 | 18 | 22 | 27 | 14.0 ~ 15.0 | 10.87 | 14.45 | 17.88 | 21.66 | 27 |
| 5 | N2 | arrd-11 RNAi | 240 | 20.17 | 0.42 | 19.35 ~ 20.99 | 17 | 21 | 25 | 27 | 40 | 19.0 ~ 19.0 | 15.29 | 19.65 | 24.05 | 26.39 | 40 |
| 5 | N2 | arrd-17 RNAi | 240 | 20.52 | 0.41 | 19.72 ~ 21.33 | 17 | 21 | 24 | 27 | 40 | 19.0 ~ 19.0 | 16.22 | 19.65 | 23.16 | 26.51 | 40 |
| 5 | N2 | arrd-19 RNAi | 240 | 19.56 | 0.41 | 18.75 ~ 20.36 | 15 | 19 | 24 | 27 | 36 | 18.0 ~ 19.0 | 14.6 | 18.7 | 23.23 | 25.9 | 36 |
| 5 | N2 | ttm-2 RNAi | 240 | 19.11 | 0.44 | 18.24 ~ 19.97 | 15 | 19 | 24 | 27 | 36 | 18.0 ~ 19.0 | 14.28 | 18.07 | 22.39 | 26.62 | 36 |
| 5 | daf-2(e1370) | control RNAi | 240 | 42.97 | 0.7 | 41.60 ~ 44.33 | 38 | 46 | 50 | 54 | 61 | 42.0 ~ 42.0 | 36.55 | 42.74 | 49.38 | 53.12 | 61 |
| 5 | daf-2(e1370) | daf-16 RNAi | 243 | 21.54 | 0.37 | 20.82 ~ 22.27 | 17 | 21 | 25 | 28 | 38 | 21.0 ~ 21.0 | 16.88 | 20.73 | 24.2 | 27.62 | 38 |
| 5 | daf-2(e1370) | arrd-11 RNAi | 210 | 42.18 | 0.72 | 40.77 ~ 43.59 | 38 | 42 | 50 | 54 | 58 | 42.0 ~ 42.0 | 35.87 | 41.78 | 49.22 | 52.53 | 58 |
| 5 | daf-2(e1370) | arrd-17 RNAi | 240 | 41.32 | 0.62 | 40.09 ~ 42.54 | 38 | 42 | 46 | 54 | 59 | - ~ - | 35.57 | 40.07 | 45.46 | 52.16 | 59 |
| 5 | daf-2(e1370) | arrd-19 RNAi | 240 | 41.1 | 0.59 | 39.94 ~ 42.26 | 38 | 42 | 46 | 50 | 59 | - ~ - | 35.72 | 40.72 | 44.68 | 49.62 | 59 |
Dataset 2.Mean lifespan and mortality rates of RNAi experiment results.
Mean lifespan, ages at different percent mortality, linear interpolation of mortality curve at specific mortality rate were calculated from pooled lifespan data of two independent lifespan experiments. Data were obtained using OASIS2 (https://sbi.postech.ac.kr/oasis2/).| Set | Condition 1 strain | Condition 1 treatment | Condition 2 strain | Condition 2 treatment | Chi square | P value | Bonferroni P value |
|---|
| 1 | N2 | control RNAi | N2 | daf-16 RNAi | 60.04 | 0 | 0 |
| 1 | N2 | control RNAi | N2 | arrd-1 RNAi | 0.69 | 0.4068 | 1 |
| 1 | N2 | control RNAi | N2 | arrd-2 RNAi | 0.08 | 0.7799 | 1 |
| 1 | N2 | control RNAi | N2 | arrd-3 RNAi | 8.09 | 0.0045 | 0.0268 |
| 1 | N2 | control RNAi | N2 | arrd-4 RNAi | 3.07 | 0.0798 | 0.4786 |
| 1 | N2 | control RNAi | N2 | arrd-5 RNAi | 0.47 | 0.4941 | 1 |
| 1 | daf-2(e1370) | control RNAi | daf-2(e1370) | daf-16 RNAi | 342.23 | 0 | 0 |
| 1 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-1 RNAi | 4.87 | 0.0274 | 0.1642 |
| 1 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-2 RNAi | 6.39 | 0.0115 | 0.0344 |
| 1 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-3 RNAi | 0 | 0.9633 | 1 |
| 1 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-4 RNAi | 2.54 | 0.1107 | 0.6644 |
| 1 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-5 RNAi | 9.05 | 0.0026 | 0.0157 |
| 2 | N2 | control RNAi | N2 | daf-16 RNAi | 60.55 | 0 | 0 |
| 2 | N2 | control RNAi | N2 | arrd-6 RNAi | 1.41 | 0.2356 | 1 |
| 2 | N2 | control RNAi | N2 | arrd-7 RNAi | 0.71 | 0.3993 | 1 |
| 2 | N2 | control RNAi | N2 | arrd-8 RNAi | 0.94 | 0.3324 | 1 |
| 2 | N2 | control RNAi | N2 | arrd-9 RNAi | 1.23 | 0.2675 | 1 |
| 2 | N2 | control RNAi | N2 | arrd-10 RNAi | 3.24 | 0.072 | 0.432 |
| 2 | daf-2(e1370) | control RNAi | daf-2(e1370) | daf-16 RNAi | 311.97 | 0 | 0 |
| 2 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-6 RNAi | 0.57 | 0.4514 | 1 |
| 2 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-7 RNAi | 0.59 | 0.443 | 1 |
| 2 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-8 RNAi | 3.79 | 0.0516 | 0.3099 |
| 2 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-9 RNAi | 3.22 | 0.0729 | 0.4376 |
| 2 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-10 RNAi | 5.82 | 0.0158 | 0.0948 |
| 3 | N2 | control RNAi | N2 | daf-16 RNAi | 158.43 | 0 | 0 |
| 3 | N2 | control RNAi | N2 | arrd-13 | 8.81 | 0.003 | 0.018 |
| 3 | N2 | control RNAi | N2 | arrd-14 | 3.81 | 0.0509 | 0.3053 |
| 3 | N2 | control RNAi | N2 | arrd-15 | 3 | 0.0831 | 0.4983 |
| 3 | N2 | control RNAi | N2 | arrd-16 | 15.5 | 0.0001 | 0.0005 |
| 3 | N2 | control RNAi | N2 | arrd-18 | 2.2 | 0.1381 | 0.8287 |
| 3 | daf-2(e1370) | control RNAi | daf-2(e1370) | daf-16 RNAi | 309.08 | 0 | 0 |
| 3 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-13 | 0.54 | 0.4607 | 1 |
| 3 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-14 | 4.09 | 0.0433 | 0.2596 |
| 3 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-15 | 0.02 | 0.8875 | 1 |
| 3 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-16 | 2.45 | 0.1174 | 0.7042 |
| 3 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-18 | 0.49 | 0.486 | 1 |
| 4 | N2 | control RNAi | N2 | daf-16 RNAi | 85.14 | 0 | 0 |
| 4 | N2 | control RNAi | N2 | arrd-23 RNAi | 7.03 | 0.008 | 0.0481 |
| 4 | N2 | control RNAi | N2 | arrd-24 RNAi | 33.81 | 0.000000006 | 0.000000036 |
| 4 | N2 | control RNAi | N2 | arrd-25 RNAi | 9.4 | 0.0022 | 0.013 |
| 4 | N2 | control RNAi | N2 | arrd-26 RNAi | 2.24 | 0.1342 | 0.8053 |
| 4 | N2 | control RNAi | N2 | arrd-28 RNAi | 1.51 | 0.2188 | 1 |
| 4 | daf-2(e1370) | control RNAi | daf-2(e1370) | daf-16 RNAi | 392.76 | 0 | 0 |
| 4 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-23 RNAi | 8.34 | 0.0039 | 0.0233 |
| 4 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-24 RNAi | 20.6 | 0.0000056 | 0.000034 |
| 4 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-25 RNAi | 24.9 | 0.00000061 | 0.0000036 |
| 4 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-26 RNAi | 11.75 | 0.0006 | 0.0037 |
| 4 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-28 RNAi | 17.71 | 0.000026 | 0.0002 |
| 5 | N2 | control RNAi | N2 | daf-16 RNAi | 67.47 | 0 | 0 |
| 5 | N2 | control RNAi | N2 | arrd-11 RNAi | 0.08 | 0.7707 | 1 |
| 5 | N2 | control RNAi | N2 | arrd-17 RNAi | 0 | 0.9914 | 1 |
| 5 | N2 | control RNAi | N2 | arrd-19 RNAi | 1.72 | 0.1891 | 0.9455 |
| 5 | N2 | control RNAi | N2 | ttm-2 RNAi | 3.22 | 0.0729 | 0.3644 |
| 5 | daf-2(e1370) | control RNAi | daf-2(e1370) | daf-16 RNAi | 368.1 | 0 | 0 |
| 5 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-11 RNAi | 1.01 | 0.3156 | 1 |
| 5 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-17 RNAi | 5.85 | 0.0156 | 0.078 |
| 5 | daf-2(e1370) | control RNAi | daf-2(e1370) | arrd-19 RNAi | 11.49 | 0.0007 | 0.0035 |
Dataset 3.Statistical analysis of lifespan data.
Statistical test results (Chi square, p value, Bonferroni p value) were calculated between ‘condition 1’ and ‘condition 2’. Test results were obtained using OASIS2 (https://sbi.postech.ac.kr/oasis2/).Discussion
The lifespan-regulatory functions of α-arrestins remain largely unexplored at the organism level. Here we showed that RNAi knockdown of individual C. elegans α-arrestins had small or no effects on lifespan in wild-type or daf-2 mutants. As knocking down each of several α-arrestins resulted in minor changes in lifespan, these C. elegans α-arrestins may play modulatory roles in longevity regulation. RNAi targeting some α-arrestins might be insufficient for causing strong lifespan phenotypes for various reasons. This may be because many of C. elegans α-arrestins are predicted to be expressed in neurons7,9,23–25, which are refractory to RNAi26–28. Lifespan assays using RNAi-hypersensitive mutants, including rrf-3(-) and eri-1(-) animals28–30, treated with α-arrestin RNAi, or using α-arrestin null mutants will help address this issue. Another possibility is that C. elegans α-arrestins may have functional redundancy, considering the large number of the α-arrestin family members in C. elegans and their sequence similarity1,7. In addition, some α-arrestins may mostly function by modulating the action of their interacting proteins6. In this case, genetic inhibition of α-arrestins may rather subtly affect the functions of their interacting partners that directly regulate physiology, such as aging and longevity, causing weak or no phenotypes. Thus, it will be interesting to test the effects of simultaneous inhibition of α-arrestins, and to identify and to functionally characterize proteins that bind C. elegans α-arrestins.
In mammals, several α-arrestins are implicated in metabolic regulation5. TXNIP (thioredoxin-interacting protein), an inhibitor of thioredoxin in mammals31–33, is a crucial negative regulator of glucose uptake34,35. ARRDC4 inhibits glucose uptake in cultured mammalian cells as well35, and ARRDC3 deficiency protects against obesity in male mice through increasing energy expenditure36. Because metabolism is closely associated with aging37, it will be interesting to test whether α-arrestins in complex metazoans play roles in aging via regulating metabolism.
Data availability
Dataset 1. Kaplan-Meier estimator of RNAi lifespan experiments. Kaplan-Meier estimate values were calculated from pooled lifespan data of two independent lifespan experiments using OASIS2 (https://sbi.postech.ac.kr/oasis2/). ‘At risk’ indicates the number of individuals at risk just prior to a specific time point. ‘S_hat’ indicates Kaplan-Meier estimate of survival function, ‘Var S_hat’ indicates variance of ‘S_hat’, and ‘SE(S)’ indicates standard error of ‘S_hat’. doi, 10.5256/f1000research.12337.d17315838
Dataset 2. Mean lifespan and mortality rates of RNAi experiment results. Mean lifespan, ages at different percent mortality, linear interpolation of mortality curve at specific mortality rate were calculated from pooled lifespan data of two independent lifespan experiments. Data were obtained using OASIS2 (https://sbi.postech.ac.kr/oasis2/). doi, 10.5256/f1000research.12337.d17315939
Dataset 3. Statistical analysis of lifespan data. Statistical test results (Chi square, p value, Bonferroni p value) were calculated between ‘condition 1’ and ‘condition 2’. Test results were obtained using OASIS2 (https://sbi.postech.ac.kr/oasis2/). doi, 10.5256/f1000research.12337.d17316040
Competing interests
No competing interests were disclosed.
Grant information
This work was supported by the Korean Government (MSIP) through the National Research Foundation of Korea (NRF) [NRF-2016R1E1A1A01941152] funded to S-J.V.L.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We thank the Lee laboratory members and Dr. Young Kwon for critical comments on the manuscript.
Supplementary material
Figure S1. RNAi knockdown of each Caenorhabditis elegans α-arrestin had minor effects on the lifespan of wild-type or daf-2(-) mutants. (A–U) Lifespan curves of wild-type (WT) and daf-2(e1370) (daf-2(-)) mutants treated with RNAi targeting daf-16 (A), which was used as a positive control, arrd-2 (B), arrd-4 (C), arrd-5 (D), arrd-6 (E), arrd-7 (F), arrd-8 (G), arrd-9 (H), arrd-10 (I), arrd-11 (J), arrd-13 (K), arrd-14 (L), arrd-15 (M), arrd-17 (N), arrd-18 (O), arrd-19 (P), arrd-23 (Q), arrd-25 (R), arrd-26 (S), arrd-28 (T) and ttm-2 (U). See Table S1 for statistical analysis and for additional repeats of lifespan data using daf-16 RNAi.
Click here to access the data.
Table S1. Lifespan data of Caenorhabditis elegans α-arrestin RNAi clones. Lifespan data within the solid lines indicate the pooled experimental datasets that are performed at the same time by two independent researchers. P values were calculated within the sets using log-rank (Mantel-Cox) method22. Percent (%) changes and p values for the lifespan data of specific α-arrestin RNAi-treated wild-type (WT) and daf-2(e1370) worms were calculated against those of control RNAi-treated WT and daf-2(e1370) worms within the sets, respectively. Percent (%) changes and p values for the lifespan data of control RNAi-treated daf-2(e1370) mutants were calculated against those of control RNAi-treated WT worms. SEM represents standard error of the mean, and 75th percentile indicates the age (days) at 75% mortality.
Click here to access the data.
Faculty Opinions recommendedReferences
- 1.
Alvarez CE:
On the origins of arrestin and rhodopsin.
BMC Evol Biol.
2008; 8: 222. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 2.
Aubry L, Guetta D, Klein G:
The arrestin fold: variations on a theme.
Curr Genomics.
2009; 10(2): 133–142. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 3.
Polekhina G, Ascher DB, Kok SF, et al.:
Structure of the N-terminal domain of human thioredoxin-interacting protein.
Acta Crystallogr D Biol Crystallogr.
2013; 69(Pt 3): 333–344. PubMed Abstract
| Publisher Full Text
- 4.
Becuwe, M, Herrador A, Haguenauer-Tsapis R, et al.:
Ubiquitin-mediated regulation of endocytosis by proteins of the arrestin family.
Biochem Res Int.
2012; 2012: 242764. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 5.
Patwari P, Lee RT:
An expanded family of arrestins regulate metabolism.
Trends Endocrinol Metab.
2012; 23(5): 216–222. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 6.
Puca L, Brou C:
Α-arrestins - new players in Notch and GPCR signaling pathways in mammals.
J Cell Sci.
2014; 127(Pt 7): 1359–1367. PubMed Abstract
| Publisher Full Text
- 7.
Hobert O:
The neuronal genome of Caenorhabditis elegans.
WormBook.
2013: 1–106. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 8.
Huffman DL, Abrami L, Sasik R, et al.:
Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins.
Proc Natl Acad Sci USA.
2004; 101(30): 10995–11000. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 9.
Jee C, Choi TW, Kalichamy K, et al.:
CNP-1 (ARRD-17), a novel substrate of calcineurin, is critical for modulation of egg-laying and locomotion in response to food and lysine sensation in Caenorhabditis elegans.
J Mol Biol.
2012; 417(3): 165–178. PubMed Abstract
| Publisher Full Text
- 10.
Tissenbaum HA:
Using C. elegans for aging research.
Invertebr Reprod Dev.
2015; 59(sup1): 59–63. Publisher Full Text
| Free Full Text
- 11.
An SWA, Artan M, Park S, et al.:
Longevity Regulation by Insulin/IGF-1 Signalling. In Ageing: Lessons from C. elegans. A. Olsen and M.S. Gill, Editors. Springer International Publishing. 2017; 63–81. Reference Source
- 12.
Altintas O, Park S, Lee SJ:
The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster.
BMB Rep.
2016; 49(2): 81–92. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 13.
Kenyon CJ:
The genetics of ageing.
Nature.
2010; 464(7288): 504–512. PubMed Abstract
| Publisher Full Text
- 14.
Lee SJ, Hwang AB, Kenyon C:
Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity.
Curr Biol.
2010; 20(23): 2131–2136. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 15.
Hwang W, Artan M, Seo M, et al.:
Inhibition of elongin C promotes longevity and protein homeostasis via HIF-1 in C. elegans.
Aging Cell.
2015; 14(6): 995–1002. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 16.
Lee D, Jeong DE, Son HG, et al.:
SREBP and MDT-15 protect C. elegans from glucose-induced accelerated aging by preventing accumulation of saturated fat.
Genes Dev.
2015; 29(23): 2490–2503. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 17.
Seo M, Seo K, Hwang W, et al.:
RNA helicase HEL-1 promotes longevity by specifically activating DAF-16/FOXO transcription factor signaling in Caenorhabditis elegans.
Proc Natl Acad Sci U S A.
2015; 112(31): E4246–E4255. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 18.
Samuelson AV, Carr CE, Ruvkun G:
Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants.
Genes Dev.
2007; 21(22): 2976–2994. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 19.
Stiernagle T:
Maintenance of C. elegans.
WormBook.
2006: 1–11. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 20.
Sievers F, Higgins DG:
Clustal omega.
Curr Protoc Bioinformatics.
2014; 48: 3.13.1–3.13.16. PubMed Abstract
| Publisher Full Text
- 21.
Huson DH, Scornavacca C:
Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks.
Syst Biol.
2012; 61(6): 1061–1067. PubMed Abstract
| Publisher Full Text
- 22.
Han, SK, Lee D, Lee H, et al.:
OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research.
Oncotarget.
2016; 7(35): 56147–56152. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 23.
Kunitomo H, Uesugi H, Kohara Y, et al.:
Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails.
Genome Biol.
2005; 6(2): R17. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 24.
Lynch AS, Briggs D, Hope IA:
Developmental expression pattern screen for genes predicted in the C. elegans genome sequencing project.
Nat Genet.
1995; 11(3): 309–313. PubMed Abstract
| Publisher Full Text
- 25.
Mounsey A, Bauer P, Hope IA:
Evidence suggesting that a fifth of annotated Caenorhabditis elegans genes may be pseudogenes.
Genome Res.
2002; 12(5): 770–775. PubMed Abstract
| Free Full Text
- 26.
Timmons L, Court DL, Fire A:
Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans.
Gene.
2001; 263(1–2): 103–112. PubMed Abstract
| Publisher Full Text
- 27.
Kamath RS, Martinez-Campos M, Zipperlen P, et al.:
Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans.
Genome Biol.
2001; 2(1): RESEARCH0002. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 28.
Kennedy S, Wang D, Ruvkun G:
A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans.
Nature.
2004; 427(6975): 645–649. PubMed Abstract
| Publisher Full Text
- 29.
Simmer F, Tijsterman M, Parrish S, et al.:
Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi.
Curr Biol.
2002; 12(15): 1317–1319. PubMed Abstract
| Publisher Full Text
- 30.
Sijen T, Fleenor J, Simmer F, et al.:
On the role of RNA amplification in dsRNA-triggered gene silencing.
Cell.
2001; 107(4): 465–476. PubMed Abstract
| Publisher Full Text
- 31.
Junn E, Han SH, Im JY, et al.:
Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function.
J Immunol.
2000; 164(12): 6287–6295. PubMed Abstract
| Publisher Full Text
- 32.
Nishiyama A, Matsui M, Iwata S, et al.:
Identification of thioredoxin-binding protein-2/vitamin D3 up-regulated protein 1 as a negative regulator of thioredoxin function and expression.
J Biol Chem.
1999; 274(31): 21645–21650. PubMed Abstract
| Publisher Full Text
- 33.
Yamanaka H, Maehira F, Oshiro M, et al.:
A possible interaction of thioredoxin with VDUP1 in HeLa cells detected in a yeast two-hybrid system.
Biochem Biophys Res Commun.
2000; 271(3): 796–800. PubMed Abstract
| Publisher Full Text
- 34.
Parikh H, Carlsson E, Chutkow WA, et al.:
TXNIP regulates peripheral glucose metabolism in humans.
PLoS Med.
2007; 4(5): e158. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 35.
Patwari P, Chutkow WA, Cummings K, et al.:
Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins.
J Biol Chem.
2009; 284(37): 24996–25003. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 36.
Patwari P, Emilsson V, Schadt EE, et al.:
The arrestin domain-containing 3 protein regulates body mass and energy expenditure.
Cell Metab.
2011; 14(5): 671–683. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 37.
Finkel T:
The metabolic regulation of aging.
Nat Med.
2015; 21(12): 1416–1423. PubMed Abstract
| Publisher Full Text
- 38.
Park S, Jung Y, An SWA, et al.:
Dataset 1 in: RNAi targeting Caenorhabditis elegans. α-arrestins marginally affects lifespan.
F1000Research.
2017. Data Source
- 39.
Park S, Jung Y, An SWA, et al.:
Dataset 2 in: RNAi targeting Caenorhabditis elegans. α-arrestins marginally affects lifespan.
F1000Research.
2017. Data Source
- 40.
Park S, Jung Y, An SWA, et al.:
Dataset 3 in: RNAi targeting Caenorhabditis elegans. α-arrestins marginally affects lifespan.
F1000Research.
2017. Data Source

Comments on this article Comments (0)