Keywords
GIT1, breast cancer, immunohistochemistry, biomarker, lymph node
GIT1, breast cancer, immunohistochemistry, biomarker, lymph node
Breast cancer is the most common type of cancer among Western women, and every year around 450,000 new cases are diagnosed in Europe alone1. Breast cancer is recognised as a very heterogeneous disease and several different subgroups have been identified on the basis of hormone receptor and Her2 status, or more recently by gene expression profiling2. Each subgroup shows different pathological, clinical and molecular characteristics, which in turn have different therapeutic options and prognostic outcome. The oestrogen-positive (ER+) breast cancer subtype is the most common of these subtypes representing around 70% of all cases. Although the majority of ER+ cases respond to anti-oestrogen treatments, some are resistant to endocrine therapies, in particular those patients with involved lymph nodes and distant metastases at time of presentation3,4. Indeed, the presence of synchronous lymph node metastasis is a strong indicator of poor prognostic outcome5. Therefore, there is a clear clinical need to identify new therapeutic targets and biomarkers for this group of patients including those that do not relapse.
It has been shown recently that advanced stage breast cancer and involved lymph nodes express high mRNA levels of G-protein-coupled receptor kinase-interacting protein 1 (GIT1)6, however, this study did not examine GIT1 protein expression, with the exception of nine tumours. As immunohistochemistry (IHC) exhibits greater potential for clinical application we decided to evaluate GIT1 immunoreactivity in an extensive cohort of ER+ breast cancer cases with synchronous lymph node (LN) involvement (n=140). We evaluated the association of GIT1 expression in the primary tumour and in the synchronous LN with clinical features and disease aggressiveness.
Primary site and lymph node metastasis tissue samples were obtained from the Pathology Department of Donostia University Hospital from 140 breast patients who were diagnosed between 2000 and 2006, with follow-up data until 2014 with a median follow-up of 8 years and 8 months. Written consent was obtained from patients for the inclusion of their samples in this study and samples were collected in accordance with the Declaration of Helsinki, and approved by local ethics committees (Comite Etico de Investigación Clínica de Euskadi (CEIC-E)). Cases were selected and re-reviewed by two experienced breast pathologists independently. Clinical data was only available for 104 of these patients (Dataset 1). All patients were women with an age range between 27 and 86 years old (median age 58 years old). All primary site tumours were ER+ and 91% of them PR+. Eighty-six percent of patients presented with invasive carcinoma of no special type (NST) and 11% with invasive lobular carcinoma (ILC). Histological grades varied from grade I (21%), grade II (55%) to grade III (15%), according to the Elston-Ellis modification of Scarff-Bloom-Richardson grading system. All patients were surgically treated either by tumorectomy or by mastectomy with axillary lymphadenectomy. All patients underwent hormone therapy with the majority of them receiving radiotherapy (90%) and adjuvant chemotherapy (75%). Table 1 summarizes the clinical and histological characteristics of patients as we previously described7.
Representative areas of high tumour load (>70%) were selected after H&E staining and two 1.5mm punch biopsies taken from both primary tumour and lymph node and arrayed non-adjacently to reduce staining bias using a Manual Tissue Arrayer MTA-1 (Beecher Instruments, USA).
Immunohistochemical (IHC) staining was carried out on 5µm slices manually using the Immunohistochemistry Accessory Kit of Bethyl Laboratories (Montgomery, USA). Slides were deparaffinized in xylene and blocked in peroxidase for 30 min. Antigen retrieval was carried out in Epitope Retrieval Buffer (Bethyl Laboratories, Montgomery, USA). Slides were blocked in BSA for 30 min and then incubated for 1h at room temperature with anti-GIT1 rabbit polyclonal antibody (1:100) (IHC-00527, Bethyl Laboratories, Montgomery, USA). After 1h incubation with secondary anti-Rabbit IHC Antibody and slides counterstained with hematoxylin.
GIT1 expression was scored in a blinded fashion by an experienced breast pathologist according to tumour cell staining intensity and categorical scores assigned as follows; 0= negative (0%); 1=1–10%; 2= 11–50%; 3= >50% (Dataset 2). Scores between non-adjacent cores were combined and categorised according to following criteria; GIT1 negative (combined score <1); moderate GIT1 expression (combined score 1–2); and high GIT1 expression (score >2). Examples of the different staining categories are shown in Figure 1 and Supplementary Figure S2. A selection of cases (n=8) were examined both as whole biopsy sections and TMAs, all scoring was concordant. Examples of whole section IHC staining with GIT1 are shown in Supplementary Figure S1 and Supplementary Figure S3.
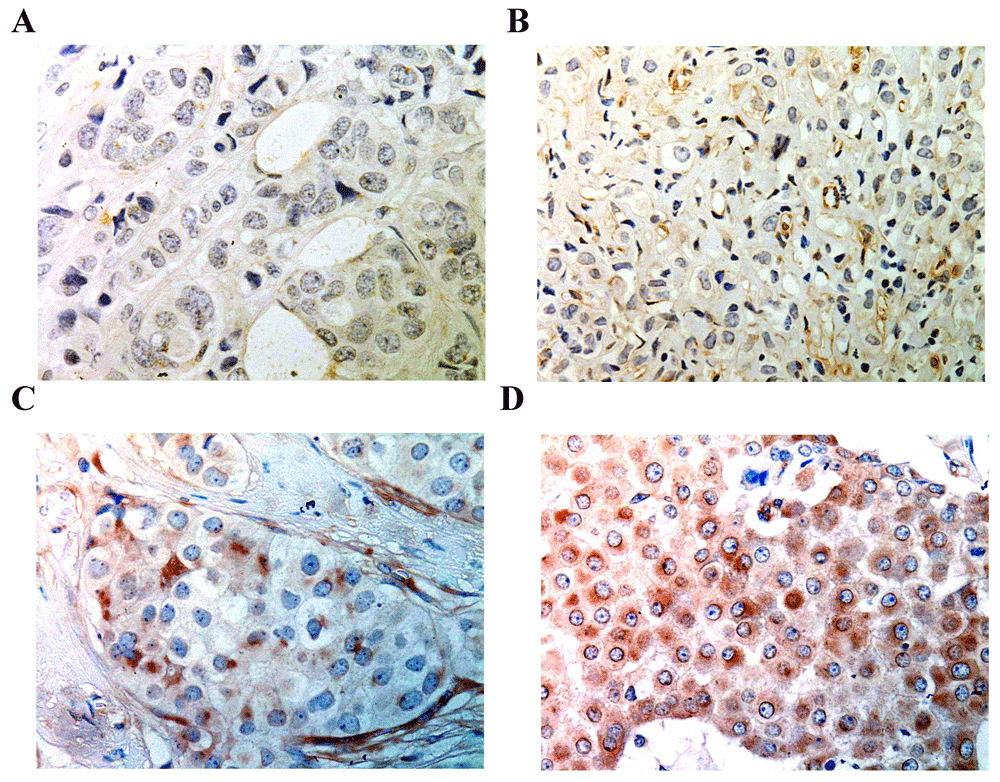
Images were enhanced from the original (Supplementary Figure S2). Representative intensity staining of GIT1 expression (primary tumour) depicting A. negative (score=0); B.weak (score=1); C. moderate (score=2); and D. strong (score=3). Magnification ×400.
GIT1 gene expression levels were analyzed using publicly available databases. For this analyses we interrogated the TCGA dataset which included 525 mixed breast cancer tumours and 22 normal breast samples (Dataset 3)8, 2000 mixed breast tumours (Dataset 4)9, 570 metastatic breast tumours (Dataset 5)10, 252 lymph-node negative breast cancer patients (Dataset 6)11, 67 triple negative breast cancer patients (Dataset 7)12, and 19 primary breast cancer and 19 brain metastasis from HER2 positive breast cancer patients (Dataset 8)13.
Chi-square statistical test was used to determine association between GIT1 expression and lymph node metastasis (Table 3). Fisher’s exact test and Chi-square test were used to associate GIT1 expression with clinicopathological features (Table 1, Table 2 and Table 4). For survival analysis, Kaplan-Meier curves and univariate Log-rank (Mantel-Cox) analysis were performed (Figure 2 and Figure 3E). Statistical analyses were performed using GraphPad Prism v5.03 (GraphPad Software, La Jolla, CA, United States). Cox regression for multivariate analysis was performed with SPSS Statistics 20 (IBM, New York, USA) (Table 5).
For public dataset analysis, expression data were analyzed by t-test when comparing 2 groups or Anova when comparing more than 2 groups (Figure 3). Data was analyzed using GraphPad Prism v5.03 (CA, United States) and R (R Foundation for Statistical Computing, Vienna, Austria).
We observed both cytoplasmic and membrane expression of GIT1 in tumour cells of varying intensities, along with some perinuclear localisation and some weaker signal in stromal cells (Figure 1). This staining pattern is consistent with that previously observed6,14.
In order to ascertain the association of tumoural GIT1 expression with clinicopathological characteristics, we scored the expression according to intensity and % positive tumour cells in each case (based on two cores) as negative, moderate and high (Figure 1). Out of the 140 primary tumour specimens, we observed high expression of GIT1 in 47 cases (34%), moderate expression in 58 cases (42%) and no GIT1 expression in 35 cases (25%). There was no significant association between GIT1 expression and the 2012 WHO defined histological subtype (i.e. invasive carcinoma of no special type (NST; n=90 (86.5%)), invasive lobular carcinoma (ILC; n=12 (11.5%)) or other). Neither were there significant associations with hormone receptor status, tumour size, number of affected lymph nodes, histological grade or the presence of vascular lymphatic infiltrate (Table 2). We observed no significant difference in the overall survival (OS) (Figure 2A), or time to recurrence in patients according to the level of GIT1 expression in these primary tumours (data not shown).
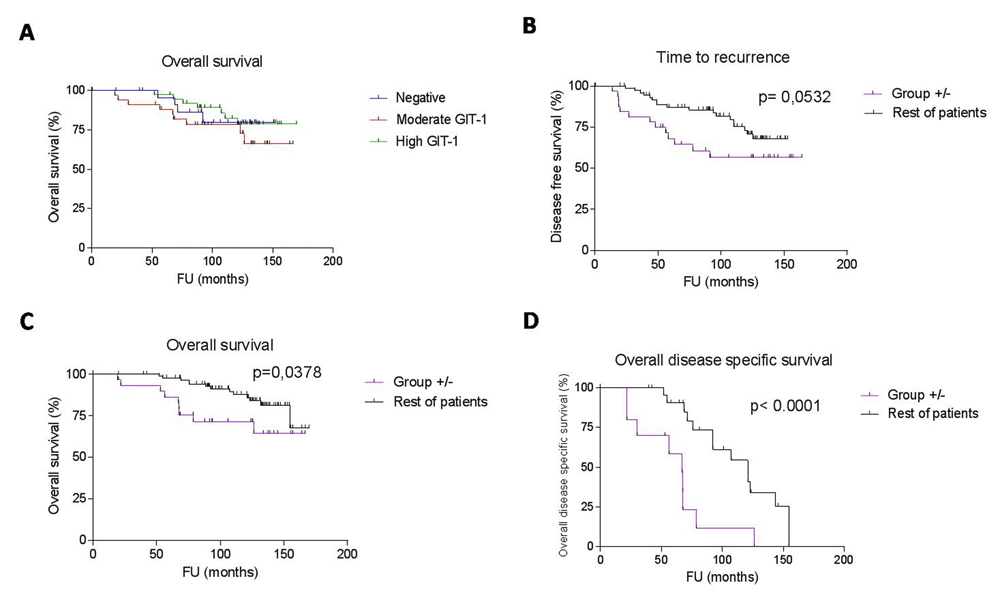
Curves were compared by univariate (log-rank) analysis. A. Cases sub-classified according to expression levels of GIT1 in the primary tumour (or LN) were not significantly different. Cases that were GIT1 +/- (n=31) had a shorter time to recurrence (B) and overall survival (C), and disease specific survival (D) than non GIT1+/- cases (n=73).
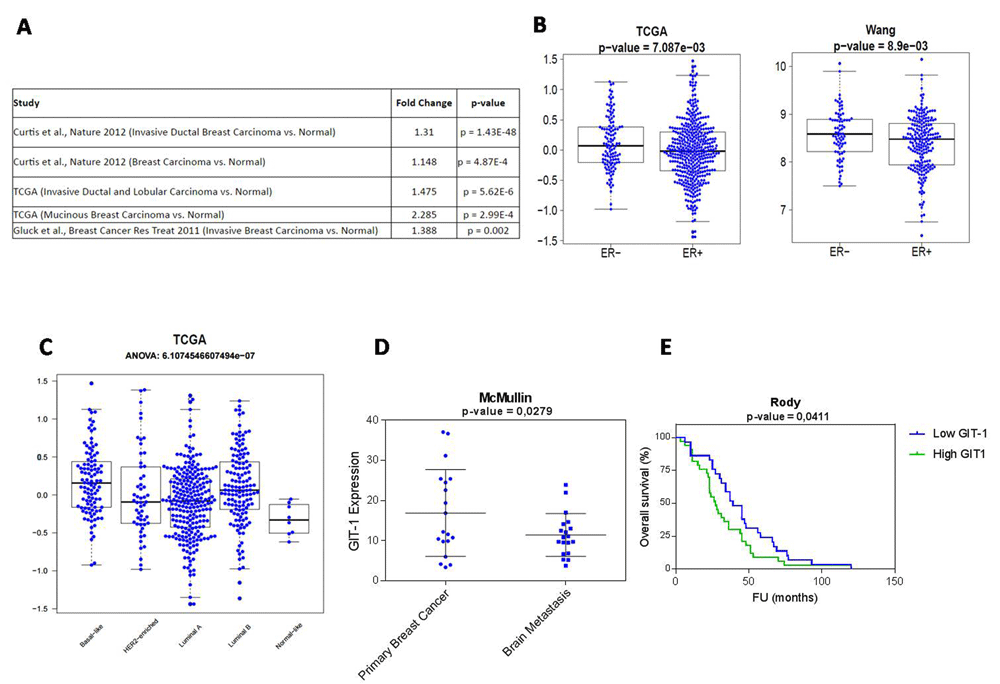
A. GIT1 expression is significantly higher in breast cancer samples compared to normal breast cancer tissue. In silico meta-analysis of five databases representing 3452 cases. B. In two independent datasets GIT1 expression was lower in ER positive compared to ER negative tumours. C. GIT1 expression is highest in the basal breast cancer subtype which represents ER-cases (ANOVA analysis). D. GIT1 expression is lower in brain metastasis than primary breast tumour sites. E. Survival analysis of triple negative breast cancer patients show low GIT1 expression is associated with poorer clinical outcome. Patients were sub-classified on the basis of median GIT1 expression levels (univariate log rank test). Unless otherwise specified analysis was carried out by independent t-test.
When we compared GIT1 expression in primary tumours with that of their counterpart lymph node metastases surprisingly we observed a significant decrease of GIT1 expression (p=0.0054, Table 3). Although both lymph node and primary biopsies showed a similar frequency of high GIT1 expression (29% vs 34% respectively), the percentage of lymph nodes with moderate GIT1 expression was lower than that of primary tumours (29% vs 41% respectively), and 43% of lymph node samples were negative for GIT1 compared with 25% of primary tumour samples (Table 3).
| GIT1 expression, n (%) | p value | |||
|---|---|---|---|---|
| Negative | Moderate | High | ||
| Tumour | 35 (25,0) | 58 (41,4) | 47 (33,6) | 0,0054 |
| Lymph node metastasis | 60 (42,9) | 40 (28,6) | 40 (28,6) | |
To explore this phenomenon further, we carried out a comparative analysis of GIT1 expression between the primary tumour site and matched synchronous lymph node metastasis. We found 64 cases (63%) with concordant GIT1 expression, either both positive for GIT1 expression (scores >1, n=45 (44%), or both negative for GIT1 expression (scores<1, n=19 (19%), and 38 cases (37%) with discordant GIT1 expression between the primary tumour and the lymph node.
As intratumoural heterogeneity has been suggested to be associated with resistance to therapy and prognostic outcome in breast cancer15, we investigated whether this spatial heterogeneity in GIT1 expression might be associated with clinical outcome (or clinicopathological characteristics) in our cohort. For this analysis we sub-classified cases into four groups: "group 0" cases were negative for GIT1 expression in both primary and lymph nodes (n=19); "group ++" cases were positive for GIT1 expression in both primary and lymph nodes (n=45); "group +/-" cases were positive for GIT1 expression in primary but not lymph nodes (i.e. loss of GIT1 expression; n=31); and "group -/+" cases were negative for GIT1 expression in primary but positive for GIT1 expression in lymph nodes (i.e. gain of GIT1 expression; n=7).
We did not detect any significant association between the aforementioned score and other clinicopathological features (Table 4). We analyzed the survival of these four groups and observed that group +/- showed a tendency towards shorter time to recurrence when compared to the rest of patients (p=0.05, hazard ratio = 2.902; Figure 2B), and that these patients had asignificantly poorer overall survival than the rest of the groups (p=0.03, hazard ratio = 2.996; Figure 2C). Furthermore, the disease specific survival of patients with +/- GIT1 expression was significantly worse than other patients (p<0.0001, hazard ratio = 7.423; Figure 2D) with a median survival time of only 67 months compared to 110 months, representing a reduction of ~40%. Comparing with other well defined prognostic indicators (histological grade, presence of distant metastasis) by multivariate analysis in our cohort, the loss of GIT1 expression in lymph nodes (+/- pattern) was an independent prognostic indicator (p=0.002; (Table 5)).
To further examine GIT1 expression in breast cancer, we analyzed gene expression levels in several publicly available gene expression datasets. This revealed that GIT1 expression was significantly higher in breast cancer (n=144) compared to non-tumoural tissue (n=14) (P=4.87 x 10-4)8–10 (Figure 3A), and was particularly pronounced when comparing only NST cases (n=1556) with healthy breast tissue (n=144) (P=1.43 x 10-48). A comparison between the two main subtypes of breast cancer (i.e. NST and ILC) in the TCGA dataset (n=525) showed differences in expression levels with healthy breast tissue (n=64) (P=5.62 x 10-6) as did the dataset of Gluck et al (n=154 vs 4 (breast carcinoma vs healthy control) (P=0.002)) (Figure 3A)10. A comparison of GIT1 expression between ER+ and ER-tumours in two independent Datasets demonstrated a significantly lower level of expression in ER+ tumours compared to ER- tumours. These comparisons were carried out on the TCGA dataset (n=601 (ER+) vs. n=179 (ER-) and the Wang dataset (n=209 (ER+) vs. n=77 (ER-)8,11 (Figure 3B). Consistent with these findings, basal tumours, which are mostly ER negative, showed higher GIT1 expression than the rest of the molecular subgroups of breast cancer in the TCGA dataset (P=6.11 x 10-7; Figure 3C).
We also looked at GIT1 expression between the primary tumour and brain metastasis in a cohort of HER2-positive (mixed ER+ and ER- cases) breast cancers (n=19)13. We observed that GIT1 expression was significantly decreased in brain metastases (P=0.0279; Figure 3D). Furthermore, when we interrogated data from a publicly available cohort of triple negative breast cancer patients (n=67)12, we found that patients with GIT1 expression below the median had significantly poorer prognosis regarding event-free interval than those with GIT1 levels above the median (P=0.0411, hazard ratio = 1.625; Figure 3E).
GIT1 is a scaffold protein that forms part of the Arf and Rho family of GTPases16. It is involved in many cellular processes including cell adhesion, migration, lamellipodia formation, cell growth and angiogenesis17–20. In addition, GIT1 can activate many signalling pathways involved in carcinogenesis such as ERK1/2, Rho, AARF or P21-activated kinase (PAK)17,21. GIT1 has been demonstrated to be over-expressed in several cancers including hepatocellular carcinoma, colon cancer, lung cancer and melanoma17,22–25.
In the current study, we ascertained the clinical relevance of GIT1 expression by IHC in a cohort of ER positive breast cancer samples with involved synchronous lymph nodes. Although it has recently been reported that GIT1 is over-expressed in lymph nodes when compared to primary breast cancer, very few cases were examined by qRT-PCR (<30) and even fewer (<10) by IHC6, the most prevalent biomarker detection technique used in clinics. Furthermore, this study did not examine the potential prognostic value of GIT1 expression or its association with distant metastasis.
We observed a significant reduction in GIT1 expression in lymph node metastasis compared to matched primary breast tumours. Furthermore, we found that over a third of cases displayed a spatial intratumoural heterogeneous pattern of GIT1 expression between the primary tumour and the lymph node, with loss of GIT1 expression in lymph nodes being more common than its gain. Heterogeneity in protein expression is a well-established phenomenon in breast cancer, particularly regarding hormone receptor status, and has been associated with prognostic outcome26. However, these studies generally report lower levels of heterogeneity (<20%) between primary and synchrononus lymph node metastases7,27, suggesting that GIT1 could be a more sensitive indicator of heterogeneity in this cancer, and hence a more powerful prognostic indicator. It should be noted that those cases with heterogeneous expression of GIT1 were not the same cases as those with heterogeneous expression of hormone receptors in this cohort. Consistent with this idea, we found that loss of GIT1 in the lymph nodes (30% of patients in this study), was indicator of poor prognosis (time to recurrence and OS) by univariate analysis and an independent indicator of prognosis by multivariate analysis. It should be noted that further analysis in independent patient cohorts within a multi-centre setting is necessary to validate these findings further.
We extended these studies to other subtypes of breast cancer by looking at cohorts from publicly available databases (n= 3452) and found that GIT1 is over-expressed in breast cancer and its expression associates inversely with ER status. Furthermore, GIT1 levels were down-regulated between primary sites and distant metastases, and that was true not only in ER+ breast cancer but also other subtypes. Despite the clear evidence shown here that GIT1 is down-regulated in both lymph node and distant metastasis in breast cancer another study reported an increase in GIT1 expression between primary tumours and lymph node metastasis6. However, it should be borne in mind that the study of Chan et al. used a much smaller cohort of patients (n=26) and moreover the hormone receptor status of these patients was not provided. As our in silico analysis (Figure 3B) suggests that GIT1 expression varies significantly with ER status, it is plausible that the subtype analysed could influence the results. Our results support the notion that, at least in ER+ breast tumours, down-regulation of GIT1 in lymph node metastases is a sign of poor prognosis.
In summary, our study has shown that the expression of GIT1 in breast cancer could serve as a useful biomarker for the management of breast cancer patients in general and for ER+/LN+ patients in particular. The mechanistic reasons behind why the loss of GIT1 in these patients is indicative of poor prognosis remains to be determined, however it is tempting to suggest that further studies could lead to better management of these patients and ultimately improve their clinical outcome.
Dataset 1: Clinical data of patient cohort. Table shows patients (numbered from 1 to 105) and their clinical features including histological subgroup, tumour size, number of affected lymph nodes, histological grade, vascular lymphatic infiltration, immunohistochemical initial status, treatment and patient follow up. 10.5256/f1000research.12393.d17543528
Dataset 2: GIT1 scoring. Table shows patients (numbered from 1 to 142) and associated primary tumour and lymph node GIT1 scoring. Categorical scores are assigned as follows according to tumour cell staining intensity; 0= negative (0%); 1=1–10%; 2= 11–50%; 3= >50%. 10.5256/f1000research.12393.d17544329
Dataset 3: Series mRNA expression matrix and clinical data information. GIT1 Expression Dataset consisting of 522 primary tumors, 3 metastatic tumors, and 22 tumor-adjacent normal samples. Data was median centered by genes. Platform: Affymetrix Human Genome U133A Array. Publicly available from https://tcga-data.nci.nih.gov/docs/publications/brca_2012/. 10.5256/f1000research.12393.d17544430
Dataset 4: Series mRNA expression matrix. Expression Dataset consisting of 2000 breast carcinoma. Platform: Affymetrix Human HT-12 V3 Array. Publicly available from http://www.cbioportal.org/study?id=brca_metabric#summary 10.5256/f1000research.12393.d17544531
Dataset 5: Series mRNA expression matrix. Expression Dataset consisting of one hundred fifty-four (154) invasive breast carcinoma samples and 4 normal breast samples. Platform: Agilent UNC Perou Lab Homo sapiens 1X44K Custom Array. Publicly available from Gluck Breast dataset (https://www.oncomine.org) 10.5256/f1000research.12393.d17544732
Dataset 6: Series mRNA expression matrix. Expression Dataset consisting of 252 lymph-node negative breast cancer samples. Platform: Affymetrix Human Genome U133A Array. Publicly available from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse2034 10.5256/f1000research.12393.d17544933
Dataset 7: Series mRNA expression matrix. Expression Dataset consisting of 67 triple negative breast cancer samples. Platform: Affymetrix Human Genome U133A Array. Publicly available from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31519 10.5256/f1000research.12393.d17545234
Dataset 8: Series mRNA expression matrix. Expression Dataset consisting of 19 HER2+ brain metastasis breast cancer samples and 19 HER2+ non-metastatic breast cancer samples. Platform: Affymetrix Human X3P Array. Publicly available from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43837 10.5256/f1000research.12393.d17545335
Written informed consent for publication of the patients' details and their images was obtained from the patients.
CHL and his research is supported by grants from the IKERBASQUE foundation for science, the Starmer-Smith memorial fund, Ministerio de Economía y Competitividad of Spanish central government and FEDER funds (PI12/00663, PIE13/00048, DTS14/00109, PI15/00275), Departamento de Desarrollo Económico y Competitividad y Departamento de Sanidad of Basque government, Asociación Española Contra el Cancer (AECC), and the Diputación Foral de Guipuzcoa (DFG). MFM acknowledges support from AECC and DFG. The work of AC is supported by FERO VIII call and the ERC starting grant (336343). IG also acknowledges support from AECC and Fundación Caja Navarra. MMC acknowledges support from IKERBASQUE foundation for science and Ministerio de Economía y Competitividad of Spanish central government. EL also acknowledges support from the Ministerio de Economía y Competitividad of Spanish central government.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Figure S1: Examples of GIT1 expression in whole tumour sections. Images were enhanced from the original (Supplementary Figure S3). Patient A demonstrates a +/- (positive primary/negative lymph node) pattern of GIT1 expression in both TMA and whole section staining. Patient B shows a -/+ (weak positive primary/positive lymph node) pattern of GIT1 expression. Magnification x40.
Click here to access the data.
Supplementary Figure S2: Example of GIT1 expression patterns found in ER+ breast cancer. Original representative intensity staining photos of GIT1 expression (primary tumour) of Figure 1. A. negative (score=0); B. weak (score=1); C. moderate (score=2); and D. strong (score=3). Magnification x400.
Click here to access the data.
Supplementary Figure S3: Examples of GIT1 expression in whole tumour sections. Original photos of Supplementary Figure S1. Patient A demonstrates a +/- (positive primary/negative lymph node) pattern of GIT1 expression in both TMA and whole section staining. Patient B shows a -/+ (weak positive primary/positive lymph node) pattern of GIT1 expression. Magnification x40.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Yoo SM, Cerione RA, Antonyak MA: The Arf-GAP and Protein Scaffold Cat1/Git1 as a Multifaceted Regulator of Cancer Progression.Small GTPases. 2017. 0 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Biochemical pharmacology of signal transduction pathways; discovered and characterized GIT1
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Cancer biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 14 Feb 18 |
read | read |
|
Version 1 30 Aug 17 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)