Keywords
Folate transporter, Eukaryotic pathogens, Drug discovery, Putative homologues
This article is included in the Neglected Tropical Diseases collection.
Folate transporter, Eukaryotic pathogens, Drug discovery, Putative homologues
A heterogeneous diversity of eukaryotic pathogens is responsible for the most economically important diseases of humans and animals1,2. As a result of underdevelopment, a lack of social infrastructure and insufficient funding of public health facilities, most of these pathogens are endemic to resource-poor countries in sub-Saharan Africa, South-East Asia and South America, where they are responsible for high morbidity and mortality1–3. Of these, parasitic protozoa form a major group, with the apicomplexans and kinetoplastid parasites represented by important members, which cause diseases such as malaria, cryptosporidiosis, toxoplasmosis, babesiosis, leishmaniasis, Human African trypanosomiasis and south American trypanosomiasis or Chagas’ disease causing most of the morbidity and mortality4,5. Other important diseases caused by protozoans include giardiasis, amoebic dysentery6,7 and trichomoniasis8. A vicious cycle of poverty and disease exists for most of these parasites with a high infection and death rate in affected populations9–11. The appreciable burden of disease caused by these parasites has been aggravated by the lack of a licensed vaccine for most of them12. Furthermore, current drugs of choice for treatment for many of the parasites have significant side effects, with the added emergence of drug resistant strains13–15. Despite the urgent demand for new therapies for control, few drugs have been developed to combat these parasites16. A major limitation to the development of new drugs is the paucity of new drug targets. There is therefore a need for discovery of novel and alternate potential chemotherapeutic targets that can help in drug development efforts for disease control16–18. A possible approach to selective antimicrobial chemotherapy has been to exploit the inhibition of unique targets, vital to the pathogen and absent in mammals17,18.
A metabolic pathway that has been exploited considerably for the development of drugs is the folate biosynthetic pathway19. Antifolate drugs target this pathway and are the most important and successful antimicrobial chemotherapies targeting a range of bacterial and eukaryotic pathogens. While most parasitic protozoa can synthesize folates from simple precursors, such as GTP, p-aminobenzoic acid (pABA) and glutamate, higher animals and humans cannot20. Additionally, a few of these parasites can also salvage folate as nutrient from their host21. These folate compounds are important for synthesis of DNA, RNA and membrane lipids and are transported via receptor-mediated or/and carrier-mediated transmembrane proteins; folate transporters20–22. Importantly, antifolate chemotherapies that target the biosynthesis and processing of folate cofactors have been effective in the chemotherapy of bacterial and protozoan parasites21. More importantly, the folate pathway has also been confirmed as being essential in some eukaryotic pathogens such as Plasmodium, trypanosomes and Leishmania19.
In addition to the folate biosynthesis pathway, proteins that mediate transport of useful nutrients such as folic acid have been identified as important chemotherapeutic drug targets18,19,23. Hence, the folate pathway, metabolites and transporters continue to be extensively studied for identification of new enzymes including transporters, which may serve as new drug targets22. Recent estimates have ascribed eight different membrane transporters to eukaryotes24.
Proteins that mediate transportation of folates have been well studied in a few parasites such as Plasmodium falciparum, Trypanosoma brucei, Leishmania donovani and Leishmania major25,26. These studies have provided information on mode of action of drugs25,27,28 in addition to studies describing mechanisms of parasite drug resistance25–32. However, folate transport proteins remain unidentified and uncharacterized in many other eukaryotic pathogens. This is despite the sequencing of the genomes of most eukaryotic pathogens, which has produced a vast wealth of data that could aid in identification of druggable pathogen-specific proteins33–39. It is therefore imperative to search and identify from these parasite genomes additional proteins such as folate transporters that may serve as novel drug targets40,41.
Therefore, in an attempt to identify and characterize targets for novel therapeutics, we report herein an extensive search of folate transporters from pathogen genomes. In addition, we investigated the evolutionary relationship of these transporters in a bid to determine similarities and differences that make them attractive drug targets. The knowledge provided may assist in the design of new antifolates for protozoan parasites.
Our experiment workflow is depicted in Figure 1. We extracted protein sequences of approximately 200 pathogens that mediate transportation or salvage of folates from Eukaryotic Pathogen Genome Database Resources (http://eupathdb.org/eupathdb/), and from the literature using a key-word search. We also searched the 200 pathogen genome sequences archived at the Eukaryotic Pathogen Genome Database Resources (http://eupathdb.org/eupathdb/). The search was for all proteins that mediate transportation or folate salvage alone or folate salvage and related compounds (such as pteridine, biopterin and methotrexate) together. This database gives public access to most sequenced emerging/re-emerging infectious pathogen genomes42. We utilized the word “folate” for search on the gene text and “folic acid” was used to confirm the hits. Hit results containing proteins annotated as folate-binding protein YgfZ, folate/pteridine transporter, folate/biopterin transporter, reduced folate carrier family protein, folate/methotrexate transporter FT1, Folate transporters alone and other folate related proteins were retrieved. The complete list of proteins extracted from Eupthadb is presented in Dataset 143. The folate transporters were classified based on type of transporter, number of transmembrane helix (TMH) and localization (either cell or mitochondrial membrane) of transporter. Gene sequences were obtained in FASTA format for transporter proteins using the sequence download tool on EupathDB (http://eupathdb.org/eupathdb/).
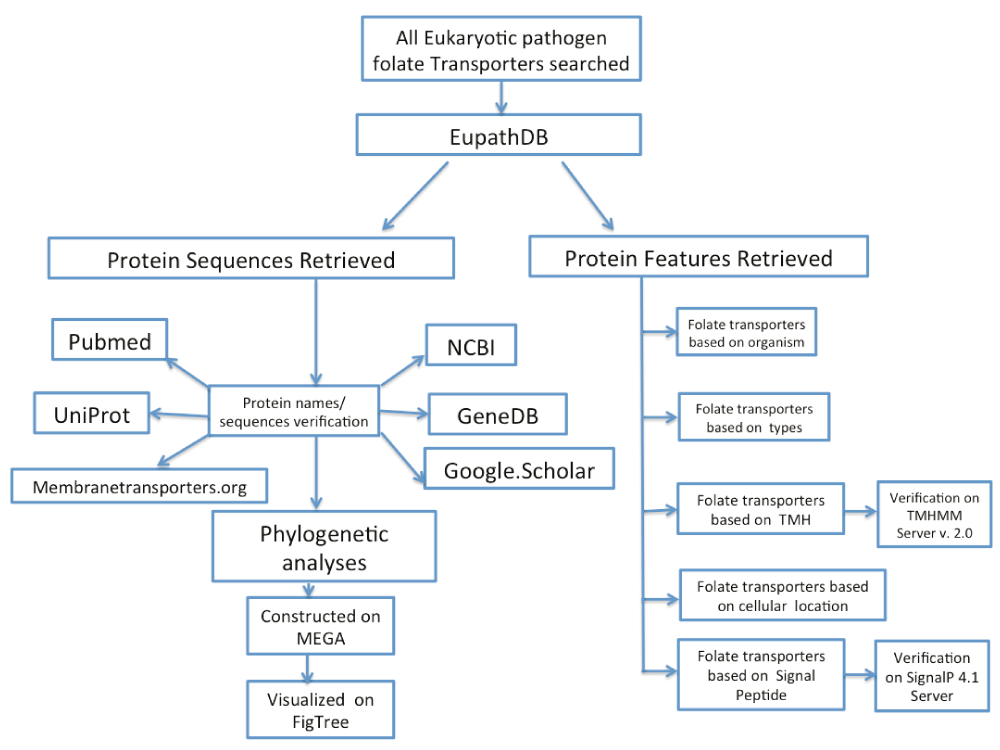
To ensure that most of the retrieved proteins had not been previously studied, we performed a literature search on PubMed (http://www.ncbi.nlm.nih.gov/pubmed/?term=) and Google Scholar (https://scholar.google.com) using the query “folate transporter + Parasite name”. The protein sequence information (Dataset 143 and Table 1) obtained from literature search was used for a BLAST search on EupathDB (http://eupathdb.org/eupathdb/), UniprotDB (http://www.uniprot.org) and GeneDB (http://www.genedb.org/Homepage). Sequence data were edited on textEdit mac version and uploaded to Molecular Evolutionary Genetics Analysis (MEGA) platform version 7.0 obtained from http://www.megasoftware.net44. The 234 sequences were aligned using muscle tools with large alignment (Max iterations = 2) selected while other settings were left at defaults. Evolutionary history was inferred using the Neighbor-Joining method45. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) was also analysed46. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the number of differences method47. While uniform rate and complete deletion was selected for substitution rates and data subset, respectively. Other parameters were at default settings. All positions containing gaps and missing data were eliminated. The newick format of the tree was exported and opened on FigTree 1.4.2 platform downloaded from http://tree.bio.ed.ac.uk/software/figtree/48. The final tree was constructed using radial tree layout. Additional analysis consisted of sub-phylogenies based on the transporter type. Since folate-binding protein YgfZ, folate/pteridine transporter, folate/biopterin transporter, putative, reduced folate carrier family protein, folate/methotrexate transporter FT1, putative folate transporters alone and others have 10, 25, 132, 2, 7, 49 and 9. So we decided to reconstruct the phylogeny based folate transporter, folate-biopterin transporter after considering the identification number, the species diversity in each category.
A total of 234 folate transporter proteins from 63 pathogens were identified from the various classes of pathogens. √, Folate transporters found from literature search; X, Folate transporters identified from this study. FTP, Folate Transporter Protein.
| S/N | Species/Strain | No. of FTP | Genera | Family | Order | Class | Phylum/Division | Kingdom | Confirmation Search (References) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | A. capsulatus G186AR | 2 | Actinobacillus | Pasteurellaceae | Pasteurellales | Gammaproteobacteria | Proteobacteria | Bacteria | X |
| 2 | A. clavatus NRRL 1 | 1 | Aspergillus | Trichocomaceae | Eurotiales | Eurotiomycetes | Ascomycota | Fungi | √69 |
| 3 | A. flavus NRRL3357 | 1 | Aspergillus | Trichocomaceae | Eurotiales | Eurotiomycetes | Ascomycota | Fungi | √70 |
| 4 | A. macrogynus ATCC 38327 | 1 | Allomyces | Blastocladiaceae | Blastocladiales | Blastocladiomycetes | Blastocladiomycota | Fungi | X |
| 5 | C. fasciculata strain Cf-Cl | 1 | Crithidia | Trypanosomatidadae | Trypanosomatida | Kinetoplastea | Euglenozoa | Eukaryota | X |
| 6 | C. immitis RS | 2 | Coccidioides | Onygenaceae | Onygenales | Eurotiomycetes | Ascomycota | Fungi | X |
| 7 | C. muris RN66 | 1 | Cryptosporidium | Cryptosporidiidae | Eucoccidiorida | Conoidasida | Apicomplexa | Eukaryota | X |
| 8 | C. neoformans var. grubii H99 | 2 | Cryptococcus | Tremellaceae | Tremellales | Tremellomycetes | Basidiomycota | Fungi | X |
| 9 | C. velia CCMP2878 | 11 | Chromera | Chromeraceae | Chromerida | Chromerida | Eukaryota | X | |
| 10 | E. acervulina Houghton | 1 | Eimeria | Eimeriidae | Eucoccidiorida | Conoidasida | Apicomplexa | Protista | X |
| 11 | E. brunetti Houghton | 2 | Eimeria | Eimeriidae | Eucoccidiorida | Conoidasida | Apicomplexa | Protista | X |
| 12 | E. cuniculi GB-M1 | 1 | Encephalitozoon | Unikaryonidae | Microsporida | Microsporea | Microsporidia | Fungi | √71,72 |
| 13 | E. maxima Weybridge | 1 | Eimeria | Eimeriidae | Eucoccidiorida | Conoidasida | Apicomplexa | Protista | X |
| 14 | E. necatrix Houghton | 1 | Eimeria | Eimeriidae | Eucoccidiorida | Conoidasida | Apicomplexa | Protista | X |
| 15 | E. praecox Houghton | 1 | Eimeria | Eimeriidae | Eucoccidiorida | Conoidasida | Apicomplexa | Protista | X |
| 16 | E. tenella strain Houghton | 1 | Eimeria | Eimeriidae | Eucoccidiorida | Conoidasida | Apicomplexa | Protista | X |
| 17 | F. graminearum PH-1 | 2 | Gibberella | Nectriaceae | Hypocreales | Sordariomycetes | Ascomycota | Animalia | X |
| 18 | H. hammondi strain H.H.34 | 3 | Hammondia | Sarcocystidae | Eucoccidiorida | Conoidasida | Apicomplexa | Eukaryota | X |
| 19 | L. braziliensis MHOM/BR/75/ M2904 | 6 | Leishmania | Trypanosomatidae | Trypanosomatida | Kinetoplastida | Euglenozoa | Eukaryota | √73 |
| 20 | L. donovani BPK282A1 | 10 | Leishmania | Trypanosomatidae | Trypanosomatida | Kinetoplastida | Euglenozoa | Eukaryota | √74,75 |
| 21 | L. infantum JPCM5 | 9 | Leishmania | Trypanosomatidae | Trypanosomatida | Kinetoplastida | Euglenozoa | Eukaryota | √76,77 |
| 22 | L. major strain Friedlin | 8 | Leishmania | Trypanosomatidae | Trypanosomatida | Kinetoplastida | Euglenozoa | Eukaryota | √78 |
| 23 | L. mexicana MHOM/GT/2001/ U1103 | 1 | Leishmania | Trypanosomatidae | Trypanosomatida | Kinetoplastida | Euglenozoa | Eukaryota | √72 |
| 24 | L. pyrrhocoris H10 | 16 | Leptomonas | Trypanosomatidae | Trypanosomatida | Kinetoplastida | Euglenozoa | Eukaryota | √79 |
| 25 | L. seymouri ATCC 30220 | 6 | Leptomonas | Trypanosomatidae | Trypanosomatida | Kinetoplastida | Euglenozoa | Eukaryota | √80 |
| 26 | M. daphniae UGP3 | 1 | Mitosporidium | – | – | – | – | – | X |
| 27 | N. bombycis CQ1 | 1 | Nosema | Nosematidae | Dissociodihaplophasida | Dihaplophasea | Microsporidia | Fungi | X |
| 28 | N. caninum Liverpool | 1 | Neospora | Sarcocystidae | Eucoccidiorida | Conoidasida | Apicomplexa | Eukaryota | X |
| 29 | N. fischeri NRRL 181 | 1 | Neosartorya | Trichocomaceae | Eurotiales | Eurotiomycetidae | Ascomycota | Fungi | X |
| 30 | N. fowleri ATCC 30863 | 2 | Naegleria | Vahlkampfiidae | Schizopyrenida | Heterolobosea | Percolozoa | Eukaryota | X |
| 31 | P. berghei ANKA | 2 | Plasmodium | Plasmodiidae | Chromatorida | Aconoidasida | Apicomplexa | Eukaryota | X |
| 32 | P. chabaudi chabaudi | 2 | Plasmodium | Plasmodiidae | Chromatorida | Aconoidasida | Apicomplexa | Eukaryota | X |
| 33 | P. falciparum 3D7 | 2 | Plasmodium | Plasmodiidae | Chromatorida | Aconoidasida | Apicomplexa | Eukaryota | √22 |
| 34 | P. falciparum IT | 2 | Plasmodium | Plasmodiidae | Chromatorida | Aconoidasida | Apicomplexa | Eukaryota | √22 |
| 35 | P. infestans T30-4 | 16 | Phytophthora | Pythiaceae | Peronosporales | Oomycota | Heterokontophyta | Eukaryota | X |
| 36 | P. knowlesi strain H | 2 | Plasmodium | Plasmodiidae | Chromatorida | Aconoidasida | Apicomplexa | Eukaryota | X |
| 37 | P. parasitica INRA-310 | 20 | Phytophthora | Pythiaceae | Peronosporales | Oomycetes | Oomycota | Fungi | X |
| 38 | P. reichenowi CDC | 2 | Plasmodium | Plasmodiidae | Chromatorida | Aconoidasida | Apicomplexa | Eukaryota | X |
| 39 | P. ultimum DAOM BR144 | 1 | Pythium | Pythiaceae | Pythiales | Oomycetes | Heterokontophyta | Chromalveolata | X |
| 40 | P. vivax Sal-1 | 2 | Plasmodium | Plasmodiidae | Chromatorida | Aconoidasida | Apicomplexa | Eukaryota | X |
| 41 | P. yoelii yoelii 17X | 2 | Plasmodium | Plasmodiidae | Chromatorida | Aconoidasida | Apicomplexa | Eukaryota | X |
| 42 | P. yoelii yoelii YM | 2 | Plasmodium | Plasmodiidae | Chromatorida | Aconoidasida | Apicomplexa | Eukaryota | X |
| 43 | S. neurona SN3 | 6 | Sarcocystis | Sarcocystidae | Eucoccidiorida | Conoidasida | Apicomplexa | Eukaryota | X |
| 44 | S. punctatus DAOM BR117 | 1 | Spizellomyces | Powellomycetaceae | Spizellomycetales | Chytridiomycetes | Chytridiomycota | Fungi | X |
| 45 | T. brucei gambiense DAL972 | 3 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | X |
| 46 | T. brucei Lister strain 427 | 9 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | √81 |
| 47 | T. brucei TREU927 | 5 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | √81 |
| 48 | T. congolense IL3000 | 2 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | X |
| 49 | T. cruzi CL Brener Esmeraldo-like | 3 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | √82 |
| 50 | T. cruzi CL Brener Non- Esmeraldo-like | 2 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | √82 |
| 51 | T. cruzi Dm28c | 2 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | √82 |
| 52 | T. cruzi marinkellei strain B7 | 6 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | √82 |
| 53 | T. cruzi Sylvio X10/1 | 2 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | √82 |
| 54 | T. equi strain WA | 4 | Theileria | Theileriidae | Piroplasmida | Aconoidasida | Apicomplexa | Eukaryota | √82 |
| 55 | T. evansi strain STIB 805 | 3 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | X |
| 56 | T. gondii GT1 | 3 | Toxoplasma | Sarcocystidae | Eucoccidiorida | Conoidasida | Apicomplexa | Eukaryota | √83 |
| 57 | T. gondii ME49 | 4 | Toxoplasma | Sarcocystidae | Eucoccidiorida | Conoidasida | Apicomplexa | Eukaryota | √83 |
| 58 | T. gondii VEG | 3 | Toxoplasma | Sarcocystidae | Eucoccidiorida | Conoidasida | Apicomplexa | Eukaryota | √83 |
| 59 | T. grayi ANR4 | 6 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | √84 |
| 60 | T. parva strain Muguga | 2 | Theileria | Theileriidae | Piroplasmida | Aconoidasida | Apicomplexa | Eukaryota | X |
| 61 | T. rangeli SC58 | 3 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | √85 |
| 62 | T. vivax Y486 | 1 | Trypanosoma | Trypanosomatidae | Trypanosomatida | Kinetoplastea | Euglenozoa | Excavata | X |
| 63 | V. brassicaformis CCMP3155 | 12 | Vitrella | Vitrellaceae | – | – | Chromerida | Eukaryota | X |
| Total | 234 | ||||||||
| Keys | |||||||||
| √ | Folate transporters found from literature search | ||||||||
| X | Folate transporters identified from this study | ||||||||
| FTP | Folate Transporter Protein |
A methodological search for folate transporters in all eukaryotic pathogen genomes we examined under EupathDB with validation via GenBank, GeneDB and Uniprot contained a total of 234 proteins (detail features of proteins are presented in Dataset 143). We identified these transporters in 28 pathogen species (containing 63 strains) cutting across 12 phyla (Table 1). The parasites with the highest number of folate transporters are Phytophthora parasitica INRA-310, P. infestans T30–4 and Leptomonas pyrrhocoris H10 with 20, 16 and 16 proteins, respectively. While Aspergillus clavatus NRRL 1, A. flavus NRRL3357, A. macrogynus ATCC 38327, Crithidia fasciculata strain Cf-Cl and others have one folate transporter protein each (Table 1). The different proteins identified to be involved in folate salvage or related molecules were folate-binding protein YgfZ, folate/pteridine transporter, folate/biopterin transporter, reduced folate carrier family protein, folate/methotrexate transporter FT1 and folate transporters having a 4%, 11%, 56%, 1%, 3% and 21% identity, respectively. Proteins that did not belong to these groups were classified as others (4%) (Figure 2A). A good number of the proteins identified had predicted transmembrane helixes with a few having none (Figure 2B). Furthermore, a number of the transporters possess signal peptides (Dataset 143), which may be required for targeting to cellular locations. Deciphering the sequence of the targeting signal may indicate its product destination.
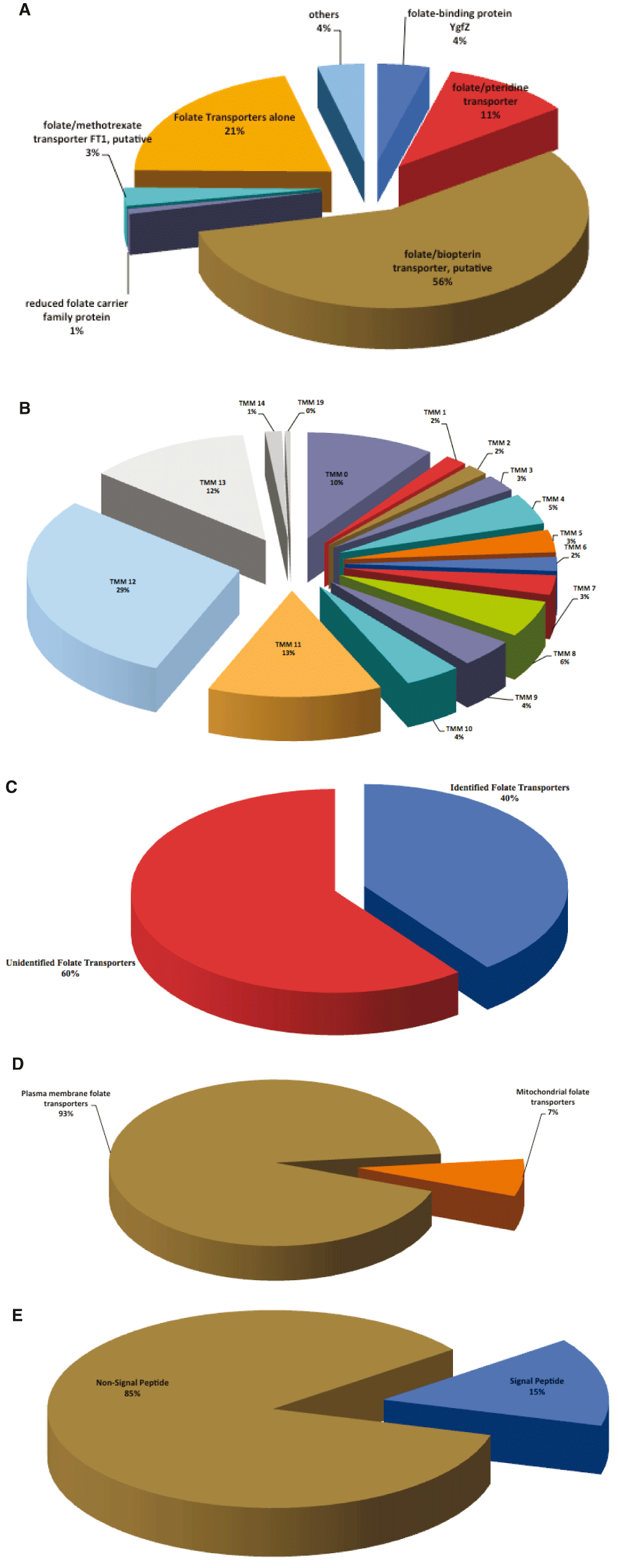
This is based on [A] Transporter Type [B] Number of TMMs [C] Novel folate transporters [D] Localization [E] Presence/absence of Signal peptide. TMM, Transmembrane Helix;
Our literature search for parasite folate transporters on PubMed and Google Scholar indicated 60% (38 out 63) of the proteins were identified for the first time as presented in Table 1 and Figure 2C, while 40% have been previously investigated. Besides, the Leishmania folate transporters we came across were not found on the EupathDB resource. We thus performed a BLAST search of Kinetoplastida on EupathDB, the returned hits were folate/biopterin transporter for L. infantum. The only Plasmodium species with results for proteins that salvage folate was P. falciparum. Our study, however, describes for the first time the presence of these transporters in other Plasmodium species. There were no transporter proteins deposited in EupathDB for P. malariae and P. ovale. However, folate transporters I and II were retrieved from our search of GeneDB for P. malariae and P. ovale curtisi, respectively.
Our analysis of folate transporters indicate the presence in Plasmodium species of two proteoforms; folate transporter I and II (Dataset 143). All Leishmania species identified possess folate/biopterin transporters and not folate transporters. Trypanosome species have both folate/pteridine and folate transporters; T. cruzi Dm28c, T. cruzi Sylvio X10/1 and T. cruzi CL Brener Esmeraldo-like, T. cruzi CL Brener Non-Esmeraldo-like and T. cruzi marinkellei strain B7 all have folate/pteridine transporter while T. brucei TREU927, T. brucei Lister strain 427, T. brucei gambiense DAL972, T. congolense IL3000 possess folate transporters. Eukaryotic parasites like Eimeria acervulina Houghton, E. brunetti Houghton, E. maxima Weybridge, E. necatrix Houghton, E. praecox Houghton, E. tenella strain Houghton and Neospora caninum Liverpool all boast folate/methotrexate transporter FT1. The folate-binding protein YgfZ was found in the fungus, Allomyces macrogynus ATCC 38327, the protist C. fasciculata strain Cf-Cl, C. immitis RS, the feline protozoon, Hammondia hammondi strain H.H.34, Sarcocystis neurona SN3, S. punctatus DAOM BR117, T. gondii GT1, T. gondii ME49, T. gondii VEG and T. brucei TREU927. Parasites such as Microsporidium daphniae UGP3 and the amoeba Naegleria fowleri ATCC 30863 possess the reduced folate carrier family protein (Figure 2D). We observed that 7% of the identified proteins are localized on the mitochondrial membrane of some pathogens such as the fungi Aspergillus clavatus NRRL 1, A. flavus NRRL3357, C. immitis RS, the yeast Cryptococcus neoformans var. grubii H99, Fusarium graminearum PH-1, A. capsulatus G186AR, Leptomonas pyrrhocoris H10, the food fungus Neosartorya fischeri NRRL 181, Phytophthora parasitica INRA-310 and P. ultimum DAOM BR144. The remaining proteins are localized on the plasma membrane (Dataset 143 and Figure 2D).
Approximately 15% (34/234) of the folate transporters identified possess signal peptides (Figure 2E) with the trypanosomes with the most signal peptides. Deductions can be made of the probable destination within the cell of any transporter by its signal peptide sequence; thus, further work may seek to decipher the sequence of the targeting signal to determine its localization. The proteins identified all have transmembrane helixes with the exception of the alveolate Chromera velia CCMP2878, apicomplexan P. berghei ANKA, S. neurona SN3, the kinetoplastid T. brucei TREU927, T. grayi ANR4 and protist Vitrella brassicaformis CCMP3155 with Gene ID’s Cvel_17766, PBANKA_0713700, SN3_01500005, Tb927.8.6480, Tgr.2739.1000 and Vbra_15327, respectively (Dataset 143).
The phylogenetic tree (Figure 3) shows the evolutionary position, history and relationship of all the folate transporters identified in this work. The type of transporter or species/strain was used for constructing phylogenic trees, with the 234 proteins identified forming two clades, a major and minor. The major clade lacked a sub-clade, while the minor clade possessed a sub-clade. All proteins identified were distributed between the two major clades; except for folate/methotrexate transporter and mitochondrial folate transporter, with the latter present on the major clade and the former on the minor clade exclusively. All the species are represented on both clades, however, V. brassicaformis CCMP3155, Plasmodium species, A. clavatus NRRL, A. flavus NRRL3357, A. macrogynus ATCC 38327, C. fasciculata strain Cf-Cl, C. immitis RS, C. immitis RS, C. muris RN66, C. neoformans var. grubii H99, C. neoformans var. grubii H99, Leishmania species, N. bombycis CQ1, N. caninum Liverpool, F. graminearum PH-1 and H. hammondi strain H.H.34 are exclusively on the major clade. There are some parasites that were identified once, as shown in Dataset 143; these are mostly in the large clade. Some of these pathogens include P. ultimum DAOM BR144, which has mitochondrial folate transporter/carrier proteins similar to Homo sapiens, E. cuniculi GB-M1, which has proteins similar to folate transporter, and S. punctatus DAOM BR117, which has folate-binding protein YgfZ. These were the only proteins of the aforementioned species identified in this work. However, M. daphniae UGP3, which had reduced folate carrier domain containing protein, was the only parasite that was found in the small clade. Improving on our phylogenetic analysis, we performed a sub-phylogenetic reconstruction (Figure 4–Figure 6) based on the substrate type of the transport proteins. After phylogenetic analysis each sub-phylogeny show a clear characterization except for folate-biopterin transporters (Figure 5), which fell in a different clade save for Leptomonas species and C. velia.

Tree was constructed using Neighbor-Joining method (Boostrap test was a 1000 replicates).
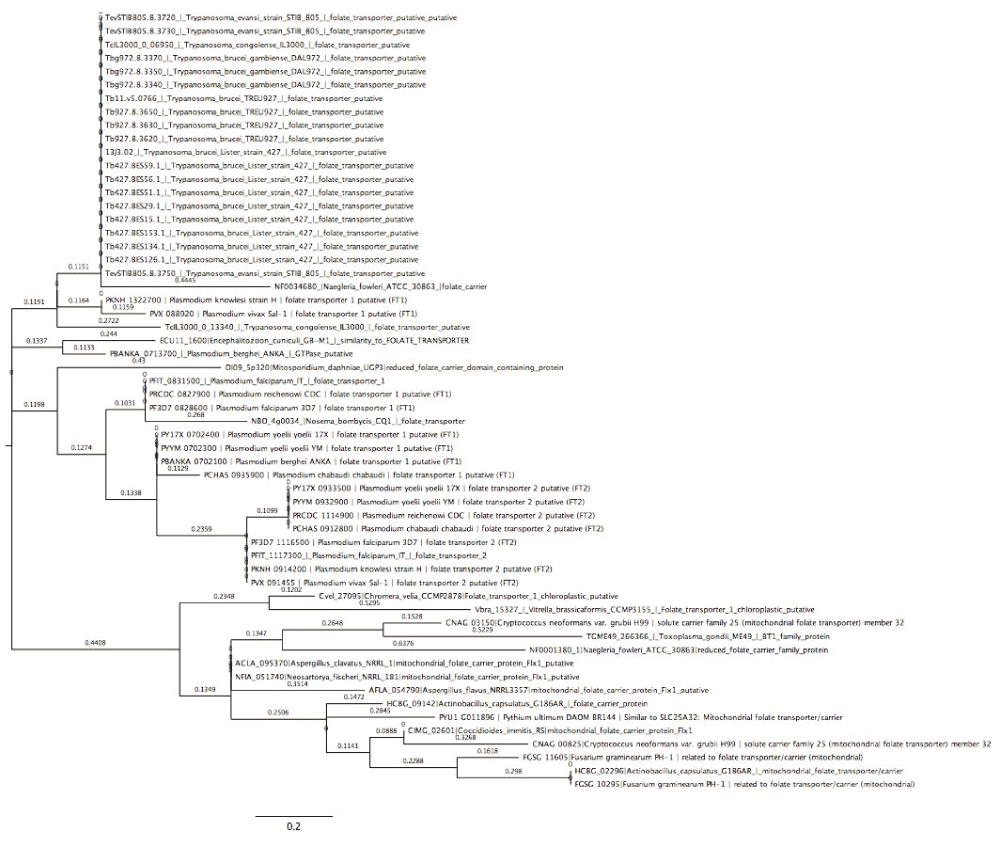
Tree was constructed using Neighbor-Joining method (Boostrap test was a 1000 replicates).
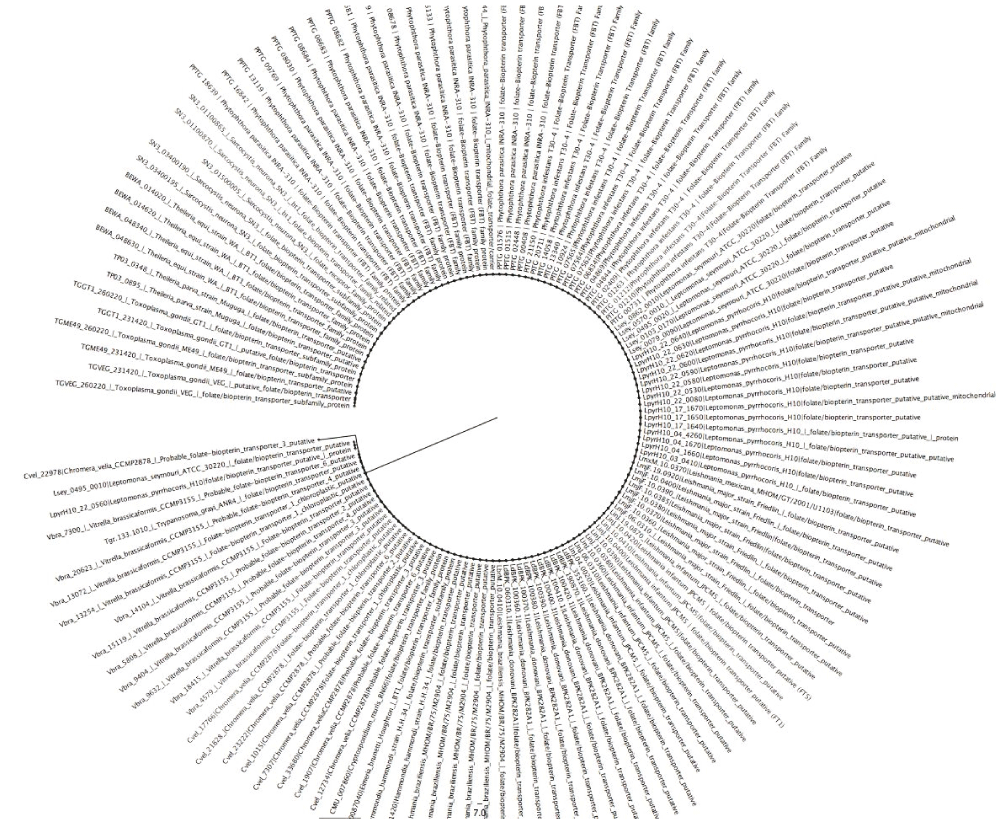
Tree was constructed using Neighbor-Joining method (Boostrap test was a 1000 replicates).
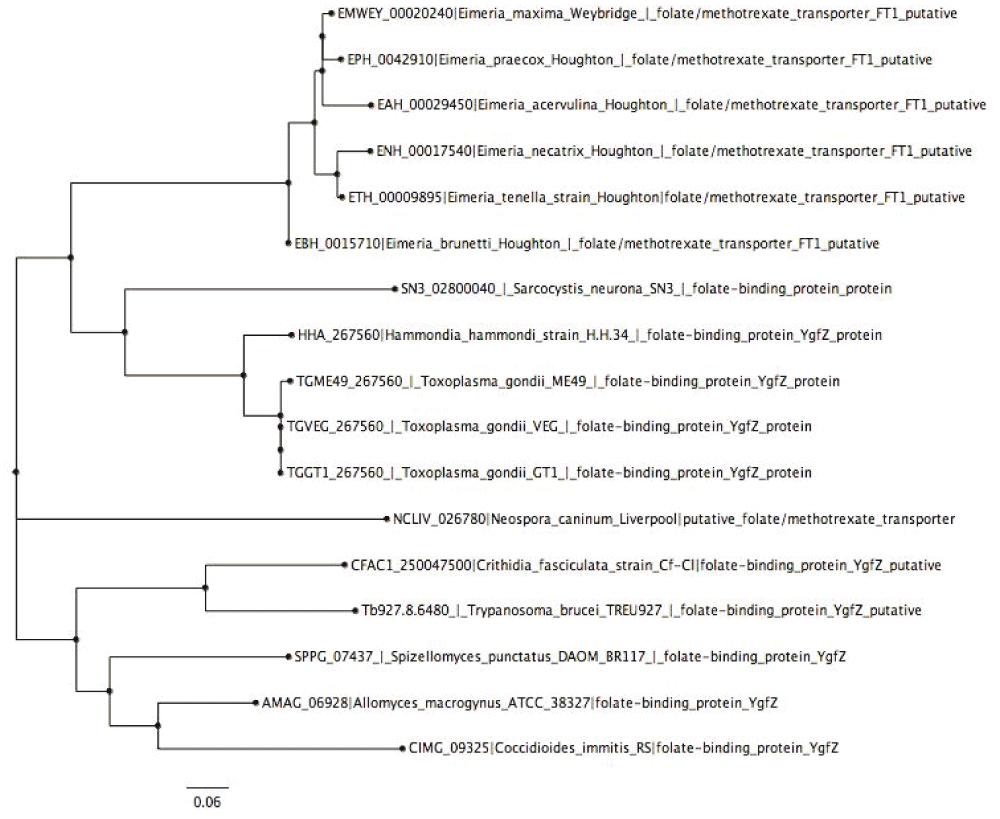
Tree was constructed using Neighbor-Joining method (Boostrap test was a 1000 replicates).
Folate transporters are important proteins involved in the salvage of folate, cofactors and related molecules in eukaryotic pathogens important for metabolism and survival in their respective hosts21. We identified proteins that could mediate the salvage of folates into cells and/or mitochondria from eukaryotic pathogen genomes in EupathDB. Many of these proteins are involved in folate biosynthesis or transport and are present in many of the eukaryotic pathogens we queried. In this study, 234 genes encoding homologues of folate salvaging proteins were identified in the genome of 64 strains, representing 28 species of eukaryotic pathogens. Some of the pathogens include P. falciparum 3D7 and IT, P. knowlesi H, P. berghei ANKA, P. chabaudi chabaudi, T. brucei Lister 427, T. brucei TREU927, T. brucei gambiense DAL972, Encephalitozoon cuniculi GB-M1. The pathogens range from bacteria through to fungi, intracellular parasites such as Plasmodium and leishmania species, to extracellular parasites such as trypanosome species. This suggests a widespread presence of the proteins cutting across a range of pathogens that infect humans and animals.
A few of the proteins we identified have been previously identified and characterized in parasites such as Plasmodium falciparum22,30, Trypanosome species26, Leishmania species and Toxoplasma gondii49. It has been estimated that over half of the drugs currently on the market target integral membrane proteins of which membrane transporters are a part, but unfortunately, these transporters have not been adequately explored as drug targets50. Folate transporters therefore represent attractive drug targets for treatment of infectious diseases. Thus their identification from other eukaryotic pathogens could open a window for novel chemotherapeutics for disease control51,52.
In Plasmodium two folate transporters have been identified, namely PfFT1 and PfT2. These transporters have been shown to mediate the salvage of folate derivatives and precursors in P. falciparum and proposed blocking of their salvage activities may improve the antimalarial efficacy of several classes of antimalarial drugs. In our work we identified folate transporters for other plasmodial species, which, like P. falciparum, may also be chemotherapeutic targets. Transport of folate in higher eukaryotes is made possible by a high affinity folate-biopterin transporters FBT or BT1 family22,30. In the trypanosomes and related kinetoplastids, a member of these transporters, the folate biopterin transporter (FBT) family of proteins was identified in Leishmania28. It is thought that MFS proteins are related to the FBT. These proteins have been characterized in a few protozoa and cyanobacteria53. Results from our study describing the presence of these transporters across several phyla corroborate results other researches, establishing the conservation of folate transport function among FBT family proteins from species from plants and protists22,53.
Malaria parasites encode transporters belonging to the organo anion transporter (OAT) folate-biopterin transporter (FBT), glycoside-pentoside-hexuronide: cation symporter (GPH), families, which are closely related to the major facilitator super-family of membrane proteins54. The inhibition of these transporters by blockers of organic anion transporters such as probenecid has been implicated in sensitization of Plasmodium resistant parasites to antifolates55,56. Thus, in Plasmodium chemotherapy, identification of folate transporters could lead to screening for compounds that interfere with folate transport and salvage for antimalarial chemotherapy22,30. We identified several types of folate transporters that have been described and functionally characterized in Leishmania with some implicated in the import of the antifolate methotrexate57,58. Thus far, only protozoan transporters in Plasmodium, Leishmania, and Trypanosoma brucei have been characterized and these are known to mediate the uptake of the vitamins folate and/or biopterin22,59,60. Thus in parasites species of medical importance folate transporter proteins may provide new targets for therapy.
We also identified folate salvaging proteins from fungi such as Coccidioides immitis and A. clavatus, fungi found in soil61–63, vegetable61 and waters in tropical and subtropical areas64. These fungi are known to occasionally become pathogenic and act as opportunistic pathogens for animals and man63. Coccidioidomycosis caused by C. immitis in association with AIDS has been known to be a fatal disease65. Treatment of acute and chronic infections with antifungals such as amphotericin B have not been adequate, hence folate transporters may present new targets in these group of pathogens. Identification was also made on pathogens such as C. fasciculata that parasitize several species of insects including mosquitoes and has been widely used to test new therapeutic strategies against parasitic infections66. As a model organism, folate transporters identified in C. fasciculata may be useful in research for developing new drugs in medically important Kinetoplasts as has been shown for other targets in this protozoan parasite67.
We noticed that P. parasitica INRA-310 and L. pyrrhocoris H10 had the highest number of folate transporters identified. Their utility as model fungal (P. parasitica) and monoxenous kinetoplast may provide models instrumental for developing new antifolates for fungal and protozoan diseases. The relatedness of these proteins across the different pathogens shows that there are two major phylogenetically distinct clades in the eukaryotic pathogens examined. The clustering of these proteins suggests that these transport proteins have highly conserved regions often required for basic cellular function or stability68–85. Thus, antifolate chemotherapic drugs that are effective against one pathogen might have some effect on others.
In summary, we have identified and classified 234 proteins after an extensive search of pathogens genome in eukaryotic pathogen resource databases, though experimental studies will be required to confirm the expression and function of these proteins in parasites. Our results show that these proteins that mediate the transportation folate are widely distributed in different pathogen species examined in various phyla. The identification of folate salvage proteins in diverse eukaryotes extend the evolutionary diversity of these proteins and suggests they might offer new possibilities for potential drug development targeting folate-salvaging routes in eukaryotic pathogens.
Dataset 1: Complete list of proteins extracted from Eupthadb and literature search, including their properties. These data are available in a .xlsx file. Doi, 10.5256/f1000research.10561.d14874243
M.O.F. and B.O. Conceptualized and Designed the study. M.O.F. and B.O. structured methodology. B.O. performed analysis. M.O.F and B.O. wrote manuscript.
B.O. was supported by a TWAS-CNPq fellowship (FP number: 3240274297).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
References
1. Woo YH, Ansari H, Otto TD, Klinger CM, et al.: Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites.Elife. 2015; 4: e06974 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 13 Jul 17 |
read | read |
|
Version 1 12 Jan 17 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)