Keywords
prenatal, fetal brain, computer-assisted radiology, Mazda, b11, cybernetics, artificial intelligence, computational visual cognition
prenatal, fetal brain, computer-assisted radiology, Mazda, b11, cybernetics, artificial intelligence, computational visual cognition
We revised the paper to improve its accuracy and clarity. The scientific content of the paper was not altered.
See the authors' detailed response to the review by Michael Hanke
See the authors' detailed response to the review by Sanjay Kalra and Abdullah Ishaque
Medicine is not an exact science but an applied interdisciplinary field1. Therefore, the time to produce physicians specialized in radiology is very long2,3. Moreover medicine is still and will still be evolving in years to come4–6. As cures are being discovered or invented, new diseases become known and mutations surface along with new variants7–10. A trainable knowledge-based system (KBS) could be an answer to the global shortage of radiologists1,11–14. Besides, there are other major obstacles, which also prevent KBS from being fully functional out of the factory. The body of medical knowledge, known to this date, is gathered and transferred through theoretical and clinical intuition as well as experience1,2,12. Then, physicians continue to expand their acquired knowledge with years of evidence-based practice15,16. On top of the challenge is the fact that some conditions may not be symptomatic during medical examination17,18. Hence, medicine is indeed a continuing learning process15,16. This is why we proposed the approach to have the observer (in this case the physicians working in the field) as the direct trainer (the programmer) of a KBS designed as a customizable, conceptual framework. In computer science, a framework is a system that implements the process of abstraction—a technique where developers make computer modeling/programming simpler to understand, use, and apply. Such a KBS should not be designed as “a hammer to drive a nail” but as an abstraction system with generic functionality, which can be changed with user written codes and customized for unlimited applications. The purpose is to enhance human-computer interaction (HCI) in medicine.
The aforementioned points are introduced especially to show the need for customization in computer-aided diagnosis (CAD). Preprogrammed CAD systems surely help radiologists and obstetricians19–21, but they could be better and more useful with some room for customization and could even improve the sharing and preservation of diagnostic innovations.
The aim of this research was to find a customizable software framework (KBS) to mathematically determine an optimal combination of texture analysis methods to differentiate anatomical structures in the developing fetal brain (i.e., regions of interest [ROI]). Why did this experiment focus on normal development of the central nervous system (CNS) rather than common conditions affecting fetal organogenesis? When considering fetal intervention to correct an anomaly in utero, the first ethical priority is actually mother safety. Prenatal well-being is clinically second, after evaluation of the benefit-risk ratio22–26. In the 1960s, fetal surgery was conceived and introduced in clinical practice27. In spite of improvements in surgical technology, the numbers of successfully treated cases of congenital defects and life expectancy of survivors are still limited28–31. The theoretical procedures are troublesome and have not been investigated enough32 . Hence, they are considered as experimental treatments32. Whether invasive or minimally invasive, fetal intervention is not the ultimate solution of this societal conundrum. It is usually reserved for cases of severe fetal anomalies28–32. In recent years, prenatal therapy is gaining popularity in religiously conservative territories particularly where abortion is prohibited33,34. Fetal intervention is recommended for fetuses with mild and non-lethal defects, in order to discourage abortion and continue childbearing. In the country where this research was carried out (i.e., Poland), abortion is, once again, nationally banned―except in cases of rape, life-threatening pregnancy and childbirth, and grave malformations33–36. Pressures are due to growing pro-life supporters demanding total anti-abortion, child-bearing at all costs, and capital punishment for illegal termination of pregnancy33–36. Nevertheless, Poland neither sanctions nor executes the outlaws of illegal abortion. Despite the absence of penalty, abortion is respectfully performed as permitted by local authority. The aforementioned dilemma justified the usage of fetal magnetic resonance imaging (fMRI) in this research. In theory, it was previously hypothesized and documented that the high electromagnetic fields (EMF) used for magnetic resonance (MR) procedures can disrupt the early stages of organogenesis. To this date, the embryotoxic, fetotoxic, and teratogenic effects of MRI are not well known. Normally, fMRI is not recommended during the first and second trimester, unless it is absolutely necessary to confirm and/or supplement the diagnosis of fetal anomalies. The legal decision to prescribe abortion to a patient sometimes requires advanced clinical investigation, accompanied by psychological counseling before and after the operations37–41. To this date, ultrasound (US) devices are preferred for obstetrical examinations. With 2D, 3D, and 4D US scans, physicians can effectively and efficiently diagnose the majority of life-threatening conditions affecting mother and fetus42–46. Therefore, ultrasonography (USG) is sufficient for diagnosis of severe malformations affecting abdominal organs. Why was fMRI prescribed? The fetal brain is where US devices struggle to produce desirable results. MRI was subsequently performed to rule out severe abnormalities in brain development, which are not visible on sonogram. MR samples came from fetuses with suspected heart and kidney defects. Disruption of organogenesis in the latter, depending on severity, might affect normal development of the brain. Specialists, structure by structure, visually investigated MR samples used in this experiment. No severe malformations were observed. In these cases, MRI studies did not add any further indication to legally fulfill the criteria to terminate pregnancy. The human visual apparatus has its limit. Unfortunately, missed diagnoses do occur. Malformations may not be apparent prior to birth. If fetal defects are suspected, bureaucracy may also restrict access to more advanced testing and healthcare. Consequently, pregnant women are deliberately forced to bear and deliver malformed babies. At the end of the day, physicians may still have to deal with the legal liability for failure to terminate pregnancy41. Unwanted fetuses may become neglected, and foundling is also a growing problem in society47–49. Congenital brain defects and its impacts on physical and cognitive development may not be detectable until after birth. Fetal outcome and mental retardation can be difficult for a physician to predict. The process requires access to better medical examination, development of more advanced tools, and further investigation. That is why the ultimate goal of this feasibility study was to gather knowledge for the practicality of a proposed project seeking to improve diagnostic accuracy and precision, by extending HCI usage in medicine. There are many CAD tools on the commercial shelves (e.g., Radiomics,50–52, Definiens Tissue Phenomics®53, and CAD4TB Diagnostic Software20,54). Sadly, they are primarily designed for pre-loaded applications but not much else.
The Research Ethics Committee of the Medical University of Lodz (MUL) approved this study. Written informed consents were obtained from all subjects, as well as perpetual licenses pertaining to copyright and ownership bundle-of-rights of the medical records for the purpose of education, research, and publication. All experiments were performed in accordance with the relevant institutional and national guidelines, regulations, licenses, and approvals from, but not limited to MUL, Lodz University of Technology (TUL), Barlicki University Hospital (BUH), Central University Hospital (CUH), Kopernik Hospital, Biegański Specialty Hospital, Polish Mother’s Memorial Hospital & Research Institute of Lodz (ICZMP), and National Health Fund (NFZ). The collective approval certificate (number RNN/213/13/KE) and the research proposal were reviewed and endorsed by the senior officers of the Research Ethics Committee.
This feasibility study was approved by the Bioethics Research Board, which regulates experiments carried out at MUL and affiliated research hospitals in Poland (permit number: RNN/213/13/KE). Due to administrative and logistic delay as well as finding volunteers and the expensive cost and availability of MRI55,56, it took us nearly five years to collect the MR samples. The nature of the study was explained to patients in Polish by attending physicians, and individual written informed consent was obtained for this research and its publication. Agreement number: 3/2011 concluded on December 6, 2011, between the experimenter (Hugues Gentillon), research supervisor (Ludomir Stefańczyk), rector of MUL (Radzisław Kordek), and the Faculty of Biomedical Science-Post-Graduate Research in Diagnostic Imaging and Radiotherapy. Consent for publication of these data was obtained. Furthermore, all data from human participants were anonymized as per consent agreement. Informed Bioethics Committee Approval Number: RNN/213/13/KE, JULY 16, 2013. If you have any questions regarding the decision please include the above number and date in your letter. Send correspondence to: THE BIOETHICS COMMITTEE OF THE MEDICAL UNIVERSITY OF LODZ Al. Kościuszki 4, 90-419 Łódź, tel. 0 785 911 596, 42 272-59-05, fax 42 272-59-07.
Though this research shares similarities with a clinical trial, it is “virtual” (i.e., it is non-interventional). Such a medical study does not meet criteria for clinical trial registration57–61. Furthermore, the investigational tools were merely used for technical exploration and to measure their feasibility in medical practice by using simulation settings. Lastly, the results were not used to alter patients’ therapeutic care and outcome57–61.
Advance notice to readers. Readers should not expect us to teach the entire science of an artificial neural network (ANN) (feedforward neural networks, recurrent neural networks, etc.) in just a manuscript. It is not possible. A full introduction is not even possible. Unfamiliar readers are expected to make an effort on their own to read and learn the basic principles of an ANN and know the basic terminology. Like a human brain, an ANN can store memories. An ANN can also judge based on stored memories and logical rules. To execute a naive trial run, it is ideal to have the ANN in a condition like “permanent global amnesia” (i.e., a phenomenon where a brain is in a state of total blackout and, thus, cannot judge based on prior memories). The KBS used in this experiment is lacking an automatic memory cleaner and optimizer. Hence, stored memories were manually deleted in the ANN for every trial run.
Computer vision. Under a reciprocal cooperation agreement between MUL and TUL, we obtained permission to test a KBS consisting of a custom version of MaZda software package62,63. It contains algorithms for data classification and visualization. How did it come to us? In 1998, a group of medical scientists, engineers, physicists, mathematicians, and others initiated the B11 Project at the European Cooperation in Science and Technology (COST). The purpose was to develop software frameworks for quantitative analysis of MR scans to improve medical diagnosis. MaZda, a Delphi/C++ computer program, was originally built in 1996 for applications in mammography. It was later used by COST to complement its MRI software modules (e.g., B11, B21). MaZda 4.6 package was the last official release (B11 version 3.3 included). In this research, we collaborated with modular programmers to further upgrade, separate, and amalgamate the functionality and compatibility of MaZda (version 5 RC HG) with other modules. In the MaZda 5 version used in this research, the new feature names were introduced to maintain compatibility with WEKA software (www.cs.waikato.ac.nz/ml/weka/; a popular package for data mining, classification, and analysis). As a result, data generated by MaZda may be used as the input to the WEKA. Also, 3D deformable models that are used for 3D volume-of-interest (VOI) generating was introduced (for details see the publication by: Piotr M. Szczypiński, Ram D. Sriram, Parupudi V.J. Sriram, D. Nageshwar Reddy, A model of deformable rings for interpretation of wireless capsule endoscopic videos, Medical Image Analysis, volume 13, issue 2, April 2009, pages 312–324). Moreover, to speed up and automate the process, some commands regarding 3D analysis were introduced. This way one can build a script (that contains commands for image loading, analysis option loading, performing analyses, and storing results) for automatic analysis of a number of 3D data. Finally, this MaZda 5 version was compiled with an Embarcadero software environment, which was not an ideal framework. Thus, the inventors decided to return to Builder C++. Currently, the latest version of MaZda being built is qMaZda (described and available at www.eletel.p.lodz.pl/pms/SoftwareQmazda.html).
Figure 1 shows the main steps of texture analysis with MaZda and B11. A key change in this custom version is the integration of MaZda with a variety of computational geometric algorithms from Qhull (e.g., VSCH_1, VSCH_2, VSCH_3, VSCH_4, and VSCH_5). For details about 1D, 2D, 3D, and 4D features see Dataset 7. VSCH network algorithms are located in the “Convex Border” menu. VSCH_1, for example, can be used to identify the strongest discriminant parameter in a large dataset.
Our trial and error experiments and texture analysis software development spanned over five years of research. All the findings shared common denominators. Quantitative brain tissue segmentation was affected by several factors, such as characteristics of fetuses (e.g., gestational age, shape, and normal/abnormal development) and the quality of fMR images (e.g., 1.5/3T, resolution, slice thickness, rotational planes, artifacts, etc.). Automatic segmentation of newborn brain MRIs have been documented in the literature64. The algorithmic contributions reported so far have achieved limited success, unfortunately. Automatic segmentation of prenatal brains is even more challenging and time consuming.
Unsupervised segmentation. Once an image was acquired in a readable format (bitmap format [BMP]), the first step was texture segmentation (i.e., partitioning an image into ROI). B11 can perform unsupervised segmentation and cluster analysis. In some instances, B11 achieved accuracy close to that of clinicians. However, unsupervised segmentation was not reliable enough for therapeutic use. Fetal brain segmentation with B11 still requires extensive expert interaction. In our observations, the key problems were maternal factors, environmental effects, growth variability, randomness of fetal movements, and its detrimental effects on image quality. Therefore, automatic segmentation was used for new insight into the possibility of improving supervised segmentation. The steps of unsupervised segmentation are relatively simple: image acquisition and run analysis. ROI and segment numbers can be manually adjusted. The drawback with B11 segmentation was its limitation to 8-bit grayscale BMP. 16-bit DICOM was converted to visually lossless BMP, by dropping the least significant bits. Note that B11 identified textures not anatomical structures. The information collected from the unsupervised trials was later used as guidance to manually estimate boundaries of anatomical structures (ROI) for the supervised trials. The preliminary trials were single-blinded (i.e., the user knew the characteristics of the ROI), and the KBS received no hints (no ROI selection). Further information was gathered on semi-automatic (unblended) segmentation by defining 4 classes (ROI): thalamus, ventricles, gray matter, and white matter. In the unsupervised mode, the KBS performed quite well when brain images were from MR examination of the same subjects and same sequences but performed poorly when they came from different subjects. The findings were likewise for the same sequences of the same patient taken at different times and MR scanner settings. The challenge was, “How do we match macroscopic characteristics with electronic recognition, regardless of MR image shadings? MaZda and B11 are not yet designed to allow the user to define semantic rules and/or import plug-ins for fully electronic recognition of anatomical structures. Object-based image analysis tools such as eCognition work consistently well for geospatial applications (e.g., identification of a river in an image)65,66. In fetal radiology, it is still a challenge to achieve consistent results with automated pattern recognition of prenatal anatomy. Programming a reliably effective system for such highly sensitive application is feasible but also time consuming. Such a task would require taking into account all known variations due to pregnancy chronology and fetal developmental, to minimize segmentation errors. MaZda and B11 were originally built for HCI rather than fully automated applications. Therefore, the best practical methodology, in this research, was for the operator to at least have prior knowledge of human embryogenesis, in order to manually and correctly identify and select fetal ROI. Often, the macroscopic appearances of many brain structures are not well differentiated in the first trimester. Hence, the selected samples were at least 20 weeks of maternal age.
Supervised segmentation. 3-tesla (3T) and 1.5-tesla (1.5T) MR sequences of fetal brain were manually segmented into 3456 ROI. The categories were predefined as follows: ventricles (class 1), thalamus (class 2), white matter (class 3), and gray matter (class 4). The selected samples did not have any brain malformations. As previously mentioned, the anomalies were in the cardiovascular and/or renal systems. The focus of this research was on normal anatomy of fetal brain. Forward processing method, also known as “supervised segmentation”67, was performed as delineated: (1) image acquisition from MR scanner, (2) selection of ROI with MaZda, (3) image normalization with MaZda, (4) feature extraction with MaZda, (5) data preprocessing with B11, and (6) texture classification with B1162,63,67. The first four steps were done with MaZda and last two steps with B11 (Figure 1). After the preliminary trials, we became interested in learning what needs to be adjusted in order to reduce misclassification of MR images. The unsupervised segmentation revealed that the KBS was very sensitive to grayscale shading, artifacts, and image thickness as well as resolution quality. Thus, we trained the KBS accordingly. 1.5T images were originally encoded as 12-bit lossless JPEG (Joint Photographic Experts Group) format and wrapped in DICOM. 1.5T images were transcoded from lossless JPEG to uncompressed DICOM. 3T images were natively uncompressed. Parameters (n = 360) were extracted with MaZda (histogram: 9; co-occurrence matrix: 220; run-length matrix: 20; gradient matrix: 5; auto-regression: 5; Haar wavelet: 28; and geometry: 73). The names of the parameters are provided in the appendix at the end of this manuscript. We assessed three automated techniques of parameter selection (i.e., Fisher selection, POE + ACC [classification error probability and average correlation coefficients], MI + PA + F [combination of mutual information, pair analysis, and Fisher selection). Further details about the mechanics and functionality of these techniques are provided in the manufacturers’ user manuals and tutorial guides68. For each technique, we also used the VSCH module in MaZda to enhance computation and visualization of geometric structures: 1) prereduction (before selection) to rule out insignificant parameters and 2) post-reduction (after selection) to identify the strongest relevant parameters. The KBS without any manual interference computed all aforementioned parts of the process automatically. Data imported (SEL [Schweitzer Engineering Laboratories] to CSV [comma-separated values]) into B11 were preprocessed with PCA, LDA, and NDA and classified by means of 1-nearest neighbor (1-NN) and ANN. In machine learning and cognitive science, 1-NN is also known as k-NN, where n = 1. It is a “lazy-learning” algorithm, in which new ROI are locally classified by getting interweaved into the closest cluster in the training set. The rest of the computation is delayed until the end of the classification process. 1-NN is implemented in B11 classifiers as well as in the preprocessing procedures in MaZda68. On the other hand, ANN is a self-organizing algorithm with hidden layers and adjustable numbers of neurons68–71. It can be used for both supervised and unsupervised classification. Neural classification algorithms are implemented in MaZda/B11. ANN training, for example, is standardized for NDA analysis68,69 (i.e., a type of feedforward ANN based on multilayer Perceptron [MLP])68. Nonlinear procedures and classifiers in B11 are MPL algorithms68,69. For optimal performance, two sets of samples are needed: one for training and another one for validation69. The pitfall with this algorithm is its sensitivity to overtraining (too strong memorization)68,69. ANN training time is shorter with standardization. For continuation, training without standardization was carried out, in spite of long processing times. ANN (one-class/n-class) and 1-NN training runs were conducted with different sequences of MR images: T2-weighted, T1-weighted, and proton density (PD)-weighted sequences. N-class training was discontinued due to repeated problems with overtraining and lack of reproducibility in F values and misclassification errors.
Despite the usage of multilevel automated selection/reduction techniques, some extracted values still did not match the controlled ROI values. Differentiating the thalamus from other thalamic nuclei and gray matter was the key problem. As a result, we manually intervened to customize and improve the extracted data. First, ROI surface areas were manually increased, in order to limit the number of parameters reporting zero and infinity values. Parameters that couldn’t be correctly computed were manually omitted in the report file. Some pre-processing procedures in both MaZda and B11 couldn’t be performed when the report file contained erroneous values. We accessed MaZda generated report files by changing the extension format from SEL to CSV and then imported them into Excel 2013 for adjustment. Parameters measured with other CAD tools can also be entered in the report files by simply using Microsoft Excel. The edited file can then be imported into B11 to perform texture classification.
Additional tests were carried out with the same ROI (i.e., thalamus, ventricle, gray matter, and white matter) to dramatically improve accuracy and precision of the KBS. It was done with a customized dataset derived from MaZda algorithms using semi-manual reduction and nearest-neighbor feature selection (see Data availability: Dataset 1–Dataset 2). The training data were used to orient the KBS to recognize what ROI had the same tissue characteristics, in spite of originating from different patients or different sequences of the same patients. The training was conducted with a combination of two built-in classification tools (i.e., NN and ANN) and four data processing techniques (i.e., RAW [read as written], PCA, LDA, and NDA). To measure the KBS sensitivity and specificity, we defined “normal” as ROI with identical tissue characteristics and defined “abnormal” as those with different tissue characteristics (Figure 2–Figure 5). Apart from noise and artifacts, we found out that the preliminary results were also affected by the planes (axial, coronal, and sagittal), which refer to the rotational planes of the spinning MR scanner in relation to the mother not the fetus. There flows the reason for the classification by rotational planes. In learning mode, we observed a consistent scoring for all the ROI. Thus the logical and semantic information provided to the KBS was effective. Statistical binary tests (also known as classification function tests) were computed in STATISTICA version 10 to assess the performance of each procedure (combination of preprocessing techniques and classifiers). In medicine, binary scores (TP, FP, TN, FN, etc.) are used to determine not just normal and abnormal characteristics but also classification property of an examination.
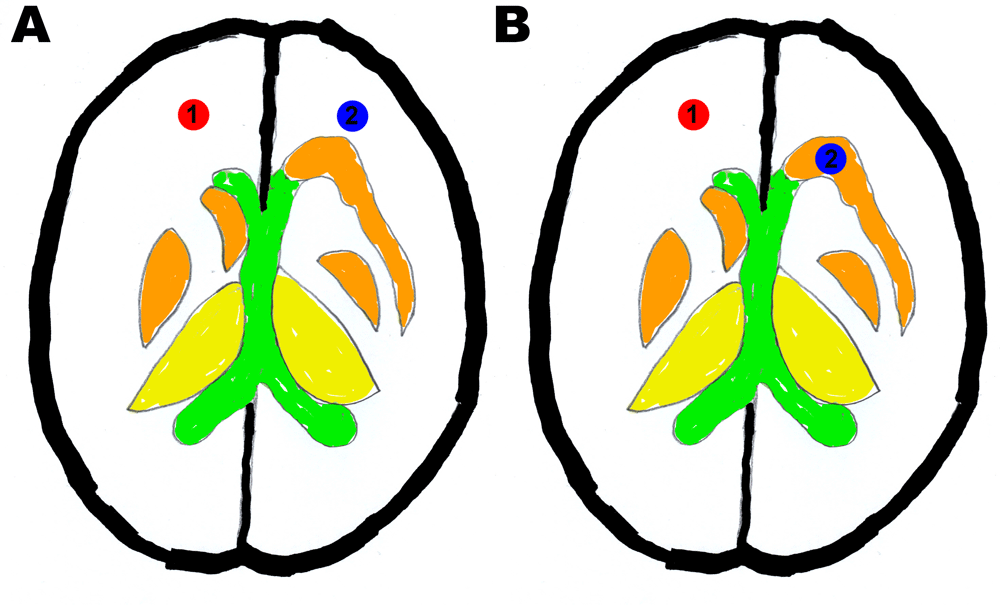
a) Specificity scoring: ROI 1 = white matter control, ROI 2 = white matter and b) Sensitivity scoring: ROI 1: white matter control, ROI 2: thalamic nucleus other than thalamus.
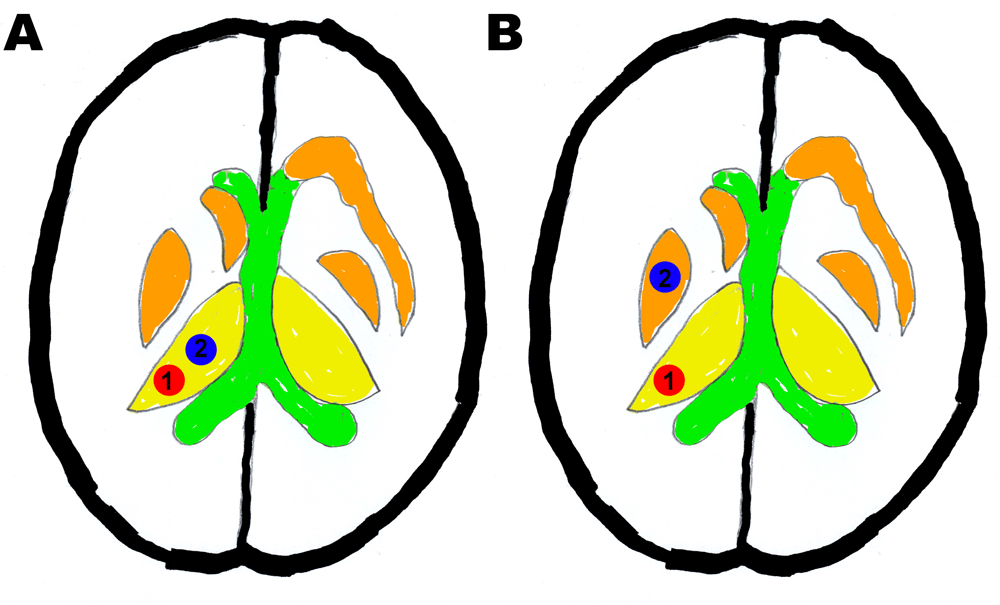
a) Specificity scoring: ROI 1 = thalamus control, ROI 2 = thalamus and b) Sensitivity scoring: ROI 1: thalamus control, ROI 2: thalamic nucleus other than thalamus.
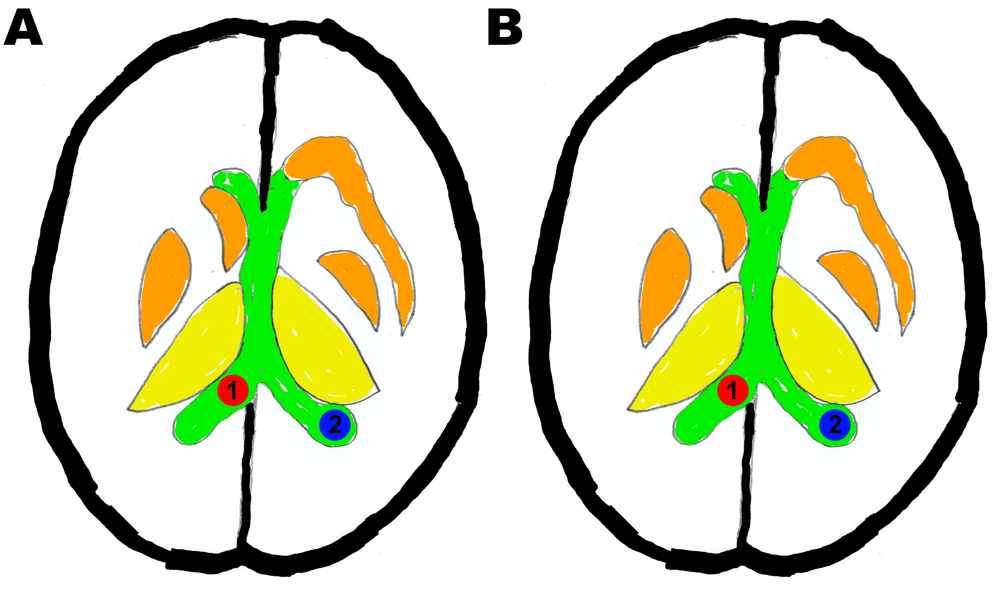
a) Specificity scoring: ROI 1 = ventricle control, ROI 2 = ventricle and b) Sensitivity scoring: ROI 1: ventricle control, ROI 2: white matter.
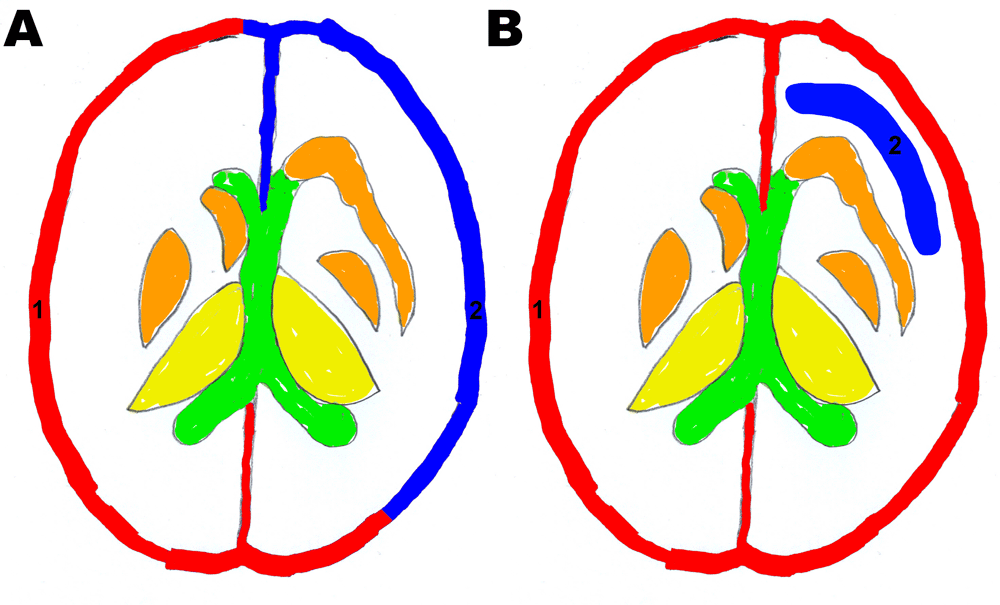
a) Specificity scoring: ROI 1 = gray matter control; ROI 2 = gray matter and b) Sensitivity scoring: ROI 1: gray matter control, ROI 2: white matter.
With the Fisher coefficient (F), we tested for the difference between ROI. It was nearly zero for ROI that were alike. Therefore, the tissue anatomy was consistently the same among the individuals in the normal ROI group. In testing mode, misclassification values, as low as 0%, were also recorded in some trials (Table 1). RAW and PCA did not respond to the training, while LDA and NDA did respond to the training. We obtained high F values, 100% sensitivity and 100% specificity, for LDA and NDA (Table 1–Table 2), which means that there was likely a real difference between the normal and the abnormal ROI.
Sn: sensitivity, Sp: specificity, CI: confidence interval, PPV: positive predictive value, NPV: negative predictive value, PLR: positive likelihood ratio, NLR: negative likelihood ratio, P: prevalence, F: Fisher coefficient, RAW: read as written, PCA: principal component analysis, LDA: linear discriminant analysis, NDA: nonlinear discriminant analysis (see Data availability: Dataset 3–Dataset 4).
Sn: sensitivity, Sp: specificity, CI: confidence interval, PPV: positive predictive value, NPV: negative predictive value, PLR: positive likelihood ratio, NLR: negative likelihood ratio, P: prevalence, F: Fisher coefficient, RAW: read as written, PCA: principal component analysis, LDA: linear discriminant analysis, NDA: nonlinear discriminant analysis (see Data availability: Dataset 5–Dataset 6).
To this date, no such research has been documented in the literature. The explanation could be derived from the difficulty of finding fMRI samples for medical research as well as the common hindrance to their availability (i.e., continuing systematic concerns over the theoretical risks of MRI usage during pregnancy), in parallel to the lack of clinical studies and trials assessing such theoretical risks72–75, plus the expensive cost of MRI examination55,56, and the scarcity of customizable CAD tools on the freeware shelves (just to list a few).
The majority of the KBS we came across were designed for technical use and not easily customizable. Such programs required paying for marketing company maintenance and for in-house developed customization service, in addition to the annual license fee. Thus, this option was not feasible for application in real-world settings where resources are often sparse (e.g., eCognition,65,66, Media Cybernetics76,77, Radiomics50–52, Definiens Tissue Phenomics®53, CAD4TB Diagnostic Software20,54, etc.).
Previous medical studies done with MaZda include inflammation, brain cancer detection, multiple sclerosis, electrophoresis, etc78–81. Herein, we defined some test samples as “abnormal” ROI. However, they were, in reality, normal tissue. Not testing directly for a common anomaly doesn’t necessarily mean that there is no real medical application. Though the tests were simulated, the research design was conceived for real-world medical applications82–84. For example, this research design could be used to detect ectopic tissue migration, neurogenesis and neuronal migration (brain function migration as a result of natural process or after injury), metaplasia, and interference with brain development. Last but not least, this simulation research followed standards used in clinical trials85.
The choice of binary classification (sensitivity/specificity) was favored over frequentist inference (p-value), because it provides more information in terms of statistical relevance to medical diagnosis, prognosis, and disease prevalence86–88. One key difference between Fisher, POE + ACC, and MI + PA + F is the number of parameters. To perform MI + PA + F, the dataset must contain at least 30 parameters strongly matching its selection-reduction criteria68. Otherwise, the KBS reported an error. In our study, it was a common occurrence when the surface area of a ROI was insufficient to extract 30 parameters meeting the MI + PA + F semantics. RAW and PCA were not so affected by the training process and, thus, remained very sensitive to minute differences in grayscale shading.
A solution to high misclassification (M) was to exclude some parameters that were very sensitive to postediting sharpness. In this research, the images were, however, processed without postediting sharpness because high M was not regarded as a problem. Instead, we used RAW and PCA as reference tests (results before the training of the KBS). On the other hand, LDA and NDA responded well to the training, and M was consistently zero.
During the study, we had to obviously clear the KBS memory several times for every trial run. We hope that the software developers will soon implement a more effective and efficient way (e.g., one click) to clear specific random access memory (RAM) without closing module(s) or without manually dumping the entire RAM or restarting the computer.
Patients gave consent to perform MRI examination, use of images in research, and for the manuscript publication. Nevertheless, this authorization was not enough, as ownership and copyright of medical records are not always exclusively attributed to patients, and such rights may not be assignable89–91. For the sake of prudence, we had to also seek institutional/research hospitals’ clearance and approval, which in turn were then subject to different administrative and logistic factors and regulations. Consequently, it took nearly five years to collect sufficient MRI samples to make this research possible.
In brief, the findings show that better results were obtained with LDA and NDA. The observed difference between the two imaging modalities was previously and repeatedly proven to be due to 3T MRI having higher resolution and being able to capture more details92–94. Lastly, LDA and NDA could be useful tests for prescreening provided ruling-in/ruling-out semantics are well defined and the KBS are well trained.
MaZda Package v5 RC HG available from: http://dx.doi.org/10.17632/dkxyrzwpzs.195.
F1000Research: Dataset 1. 1.5T data, 10.5256/f1000research.10401.d14678296.
F1000Research: Dataset 2. 3T data, 10.5256/f1000research.10401.d14678397.
F1000Research: Dataset 3. 1.5T, 10.5256/f1000research.10401.d14678498.
F1000Research: Dataset 4. 3T, 10.5256/f1000research.10401.d14678599.
F1000Research: Dataset 5. 1.5T, 10.5256/f1000research.10401.d146786100.
F1000Research: Dataset 6. 3T, 10.5256/f1000research.10401.d146787101.
F1000Research: Dataset 7. Parameter list, 10.5256/f1000research.10401.d146788102.
HG: experimenter, research designer, and writer; LS: supervisor, sample provider, and clinical feedback; MRL: coordinator, sample provider, and clinical feedback; MS: software maker, updates, and technical feedback.
Medical University of Lodz & Polish Research Committee and affiliated institutions and hospitals; Self-funded; MRI cost was covered by Polish National Health Fund, grants and financial aid from Swedish Ministry of Education and Research/Centrala Studiestödsnämnden and from U.S. Department of Education.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors gratefully acknowledge Rafal Pawliczak and MUL staff for research coordination, logistics, and administration; Paweł Liberski and MUL Neuropathology Department for counsels with funds to cover MRI expenses; Ludomir Stefańczyk and Barlicki Hospital staff for sample supply and clinical feedback; Maria Respondek-Liberska and Matki Polki Hospital for sample supply and clinical feedback; Tadeusz Biegański, ICZMP Director; Michał Strzelecki and TUL staff for providing MaZda software and technical feedback.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Neuroimaging
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 11 Sep 17 |
read | |
|
Version 1 12 Jan 17 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
1. In this feasibility study, we proposed that "the observer (in this case the ... Continue reading In version 2, we did not change the scientific content of this paper for the following reasons:
1. In this feasibility study, we proposed that "the observer (in this case the physicians working in the field) as the direct trainer (the programmer) of a KBS designed as a customizable, conceptual framework." In the introduction, we further reiterate (re-emphasize) that "the ultimate goal of this feasibility study was to gather knowledge for the practicality of a proposed project seeking to improve diagnostic accuracy and precision, by extending HCI usage in medicine." Last but not least, we clearly stated that this research is not a clinical trial and registration was not required.
2. We feel that the reviewers were not so familiar with the process of clinical trials (esp. the various stages). In this manuscript, we cannot teach everything about clinical trials, just as we cannot also teach everything about the science of artificial neural network.
3. Herein we are taking the opportunity to further explain the purpose of a "feasibility study". A feasibility study is the earliest stage of clinical trials. Its focus is on pre-clinical development, pre-clinical studies and/or non-clinical studies. A feasibility study itself also has different parts. This paper focuses on the initial phase. Herein we are not testing a drug but a "robotware" -- i.e. a software, a piece of a potentially larger A.I. system.
4. We appreciate all the referees' critics and suggestions. Unfortunately, we had to strongly rebut the negative comments because they were influenced by misinterpretations and earlier negative comments. This manuscript is not, per se, a report of a clinical research but a feasibility study. In terms of clinical trials, a feasibility study may contain pre-clinical and/or non-clinical components. In the future, another experiment may be done to extensively test for pre-clinical components, such as biocompatibility and safety. Before we even get there, we had to carry out this preliminary research to investigate the non-clinical aspects of the project. That was the intended interpretation being sought from the readers.
5. We must not confuse scientific medicine with clinical medicine. Scientific medicine is hypothetical and experimental. Clinical medicine directly deals with applications, diagnosis, treatment, prognosis, etc. At this stage, the research focus is primarily on the non-clinical aspects of the project. As a result, we decided to omit unnecessary clinical information from this publication.
6. Why did we deviate from the philosophy of testing pathological versus non-pathological? The reason is computational semiotics. Unlike adolescents and adults, children naturally learn the architecture of normally spoken language without conscious learning. They perfectly imitate pronunciation and figure out the rules by themselves. In this experiment, our training approach was somewhat based on the aforementioned phenomenon. At this stage, we do not need various types of cancer tissues to train the software to recognize what is a normal texture for thalamus, for example.
7. It is important to also understand that "abnormal" does not necessarily mean pathological. In terms of neuroplasticity, our testing methods do have a clinical relevance.
We invite referees to read this paper with an open mind. Here are some inspirational words of wisdom from Charles F. Kettering : "People are very open-minded about new things… as long as they're exactly like the old ones." If we have to please publishing expectations, then it is not a feasibility research.
1. In this feasibility study, we proposed that "the observer (in this case the physicians working in the field) as the direct trainer (the programmer) of a KBS designed as a customizable, conceptual framework." In the introduction, we further reiterate (re-emphasize) that "the ultimate goal of this feasibility study was to gather knowledge for the practicality of a proposed project seeking to improve diagnostic accuracy and precision, by extending HCI usage in medicine." Last but not least, we clearly stated that this research is not a clinical trial and registration was not required.
2. We feel that the reviewers were not so familiar with the process of clinical trials (esp. the various stages). In this manuscript, we cannot teach everything about clinical trials, just as we cannot also teach everything about the science of artificial neural network.
3. Herein we are taking the opportunity to further explain the purpose of a "feasibility study". A feasibility study is the earliest stage of clinical trials. Its focus is on pre-clinical development, pre-clinical studies and/or non-clinical studies. A feasibility study itself also has different parts. This paper focuses on the initial phase. Herein we are not testing a drug but a "robotware" -- i.e. a software, a piece of a potentially larger A.I. system.
4. We appreciate all the referees' critics and suggestions. Unfortunately, we had to strongly rebut the negative comments because they were influenced by misinterpretations and earlier negative comments. This manuscript is not, per se, a report of a clinical research but a feasibility study. In terms of clinical trials, a feasibility study may contain pre-clinical and/or non-clinical components. In the future, another experiment may be done to extensively test for pre-clinical components, such as biocompatibility and safety. Before we even get there, we had to carry out this preliminary research to investigate the non-clinical aspects of the project. That was the intended interpretation being sought from the readers.
5. We must not confuse scientific medicine with clinical medicine. Scientific medicine is hypothetical and experimental. Clinical medicine directly deals with applications, diagnosis, treatment, prognosis, etc. At this stage, the research focus is primarily on the non-clinical aspects of the project. As a result, we decided to omit unnecessary clinical information from this publication.
6. Why did we deviate from the philosophy of testing pathological versus non-pathological? The reason is computational semiotics. Unlike adolescents and adults, children naturally learn the architecture of normally spoken language without conscious learning. They perfectly imitate pronunciation and figure out the rules by themselves. In this experiment, our training approach was somewhat based on the aforementioned phenomenon. At this stage, we do not need various types of cancer tissues to train the software to recognize what is a normal texture for thalamus, for example.
7. It is important to also understand that "abnormal" does not necessarily mean pathological. In terms of neuroplasticity, our testing methods do have a clinical relevance.
We invite referees to read this paper with an open mind. Here are some inspirational words of wisdom from Charles F. Kettering : "People are very open-minded about new things… as long as they're exactly like the old ones." If we have to please publishing expectations, then it is not a feasibility research.