Keywords
Amino acid, Glutamyl semialdehyde, Oxidative stress, Protein carbonylation, Protein engineering, Protein oxidation, Reactive oxygen species
Amino acid, Glutamyl semialdehyde, Oxidative stress, Protein carbonylation, Protein engineering, Protein oxidation, Reactive oxygen species
Reactive oxygen species (ROS) are produced through the electron reduction of molecular oxygen and include superoxide anion radicals, hydrogen peroxide (H2O2), and hydroxyl radicals (Freeman & Crapo, 1982; Halliwell & Gutteridge, 2007). ROS have been implicated in the pathogenesis of various diseases (Freeman & Crapo, 1982; Halliwell & Gutteridge, 2007), as well as in the aging process (Harman, 1956). One electron reduction of molecular oxygen produces superoxide, which in turn reacts with each other to produce H2O2 and reduces cellular iron ions. Reduced iron donates an electron to H2O2 and produces highly reactive hydroxyl radicals. Hydroxyl radicals in turn react with virtually all biological molecules, including DNA, proteins, lipids and small molecules, damaging the biological system (Freeman & Crapo, 1982; Halliwell & Gutteridge, 2007).
One important event that occurs in response to the metal (iron)-catalyzed oxidation process is the formation of carbonyls in the protein structure. Protein carbonylation has been shown to be increased in various diseases and in aging (Berlett & Stadtman, 1997; Levine & Stadtman, 2001; Levine, 2002; Stadtman et al., 1988). Protein carbonylation occurs in response to the iron-catalyzed, H2O2-dependent oxidation of amino acid side chains (Stadtman, 1990; Suzuki et al., 2010). Protein carbonylation inactivates protein functions and marks damaged proteins for proteasome-dependent degradation (Grune et al., 1997; Levine, 1989). While carbonylated proteins are believed not to undergo electron reduction, we previously discovered the protein de-carbonylation mechanism, in which carbonyl groups can be eliminated without proteins being degraded (Wong et al., 2008). Major amino acid residues that are susceptible to iron-catalyzed oxidation include proline and arginine, both of which are oxidized to become glutamyl semialdehyde, which contains a carbonyl group (Amici et al., 1989). Glutamyl semialdehyde is further oxidized into glutamic acid (Figure 1).
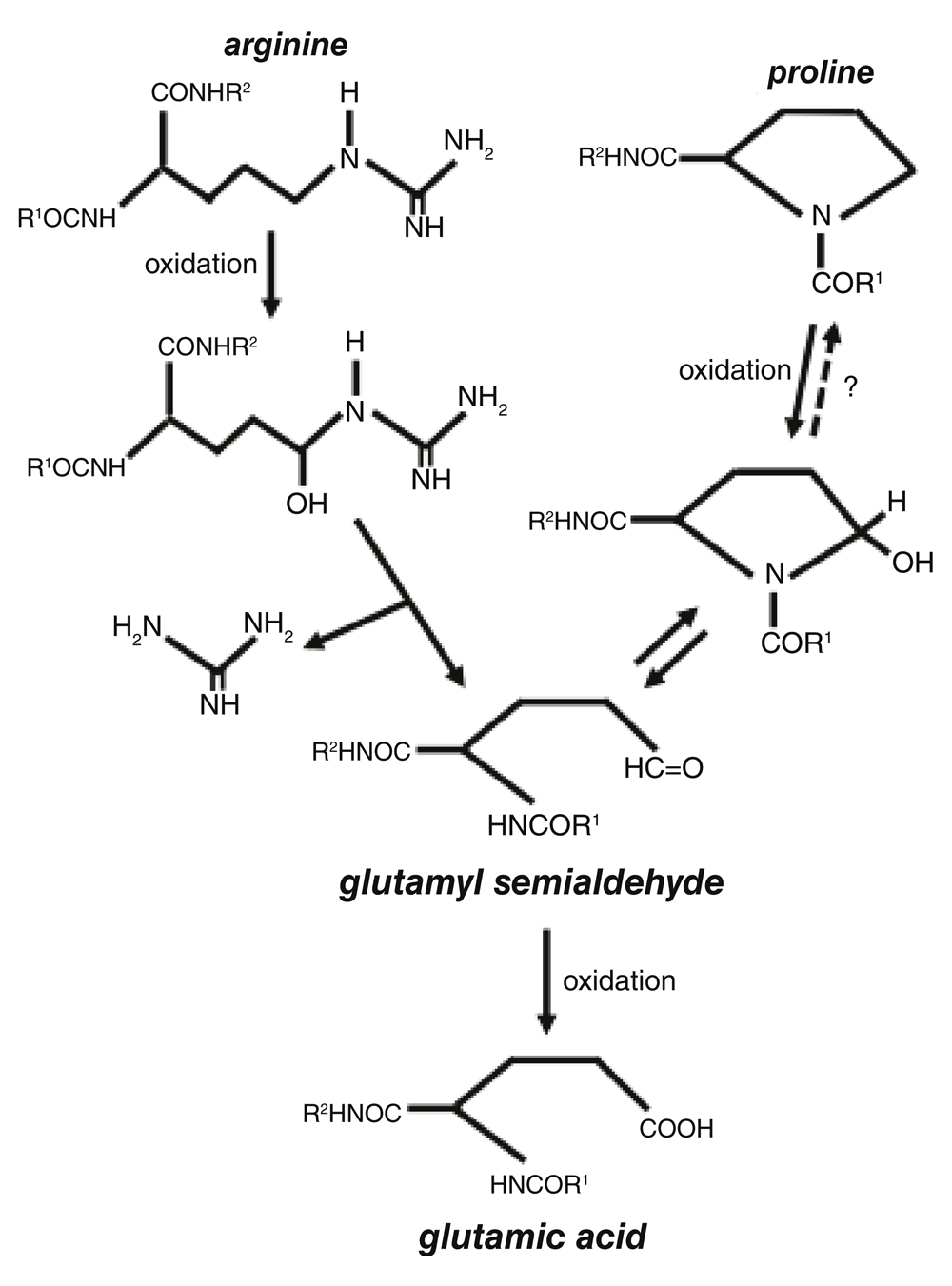
Glutamyl semialdehyde is further oxidized into glutamic acid.
We previously demonstrated the role of protein carbonylation in ligand/receptor-mediated cell signaling (Wong et al., 2008). We further noted that the kinetics of ligand-mediated protein carbonylation is transient. Typically, in cultured cells, ligands activate the carbonylation of various proteins within 10 min and the activated protein carbonylation reverts to baseline by 30 min. These results suggest that there is a mechanism for the elimination of the formed carbonyls. We named this process “de-carbonylation” (Wong et al., 2008). To understand the mechanism of de-carbonylation, we tested the hypothesis that protein carbonyls may be reduced. We found that the addition of reductants to rat heart homogenates resulted in a decrease in the protein carbonyl content (Wong et al., 2013). By contrast, reductants had no effect on the carbonyl content in purified proteins, suggesting that protein carbonyls are not reduced in the absence of other cellular components. From these results, we hypothesized that cells contain catalysts for the reduction of protein carbonyls. This hypothesis is supported by our results demonstrating that the heating of heart homogenates to inactivate cellular enzymes inhibits the decrease in protein carbonyls in vitro, and that knocking down glutaredoxin 1 in the cells inhibits protein de-carbonylation (Wong et al., 2013). We used two-dimensional gel electrophoresis and mass spectrometry to identify proteins that can be de-carbonylated and found that peroxiredoxin 6 (Prx6) is one such protein (Wong et al., 2013).
Since both arginine and proline residues can be oxidized to form glutamyl semialdehyde that can further be oxidized to form glutamic acid, we speculated that arginine, proline, and glutamic acid residues may be interchangeable in the biological system, in a process that resembles site-directed mutagenesis. This article reports that the proline residue 45 of the human Prx6 protein molecule can be converted into glutamic acid in cells, indeed demonstrating the existence of a naturally occurring site-directed mutagenesis/protein engineering-like process that may be regulated by ROS.
Human pulmonary artery smooth muscle cells (ScienCell Research Laboratories, Carlsbad, CA, USA) grown in 10 cm dishes were serum-starved overnight with 10 ml of 0.01% fetal bovine serum-containing Dulbecco’s Modified Eagle’s medium (Mediatech, Inc., Manassas, VA, USA) for cell signaling studies. To prepare lysates, the cells were washed with phosphate buffered saline and solubilized with 1 ml of 50 mM Hepes solution (pH 7.4) containing 1% (v/v) Triton X-100, 4 mM EDTA, 1 mM sodium fluoride, 0.1 mM sodium orthovanadate, 1 mM tetrasodium pyrophosphate, 2 mM PMSF, 10 µg/mL leupeptin, and 10 µg/mL aprotinin. Cell lysates (1 ml) were immunoprecipitated with the rabbit polyclonal anti-Prx6 antibody (Sigma-Aldrich, St. Louis, MO, USA; Catalogue # P0058; 5 µg) and SureBeads Protein G Magnetic Beads (Bio-Rad Bio-Rad Laboratories, Hercules, CA, USA; 1 mg) for 1 h at room temperature.
Immunoprecipitation samples were processed with trypsin digestion (12.5 ng/µl) followed by a C18 Zip-tip clean-up (EMD Millipore, Billerica, MA, USA). Tryptic peptide samples were reconstituted in 20 µl of 0.1% formic acid before nanospray liquid chromatography/mass spectrometry/mass spectrometry (LC/MS/MS) analysis was performed.
The tryptic peptides mixture from each sample was analyzed using a Thermo Scientific Q-Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Electron, Bremen, Germany) equipped with a Thermo Dionex UltiMate 3000 RSLCnano System (Thermo Dionex, Sunnyvale, CA, USA). Tryptic peptide samples were loaded onto a peptide trap cartridge at a flow rate of 5 μl/min. The trapped peptides were eluted onto a reversed-phase 20-cm C18 PicoFrit column (New Objective, Woburn, MA, USA) using a linear gradient of acetonitrile (3–36%) in 0.1% formic acid. The elution duration was 60 min at a flow rate of 0.3 μl/min. Eluted peptides from the PicoFrit column were ionized and sprayed into the mass spectrometer using a Nanospray Flex Ion Source ES071 (Thermo Scientific, Waltham, MA, USA) under the following settings: spray voltage 1.6 kV and capillary temperature 250°C. The Q Exactive instrument was operated in the data-dependent mode to automatically switch between full scan MS and MS/MS acquisition. Survey full scan MS spectra (m/z 300−2,000) were acquired in the Orbitrap with 70,000 resolution (m/z 200) after the accumulation of ions to a 3 × 106 target value based on predictive AGC from the previous full scan. Dynamic exclusion was set to 20 s. The 15 most intense multiply charged ions (z ≥ 2) were sequentially isolated and fragmented in the Axial Higher Energy Collision-induced Dissociation (HCD) cell using normalized HCD collision energy at 25% with an AGC target of 1e5 and a maximum injection time of 100 ms at 17,500 resolution. Two independent MS analyses in triplicate (a total of six cell samples) were performed.
The raw MS files were analyzed using the Thermo Proteome Discoverer 1.4.1 platform (Thermo Scientific, Bremen, Germany) for peptide identification and protein assembly. The raw data files were searched against the human protein sequence database obtained from the NCBI website (https://www.ncbi.nlm.nih.gov) using the Proteome Discoverer software based on the SEQUEST algorithm. The carbamidomethylation of cysteines was set as a fixed modification, and Oxidation and Deamidation Q/N-deamidated (+0.98402 Da), and Pro>Glu (+31.990 Da) were set as dynamic modifications. The minimum peptide length was specified to be five amino acids. The precursor mass tolerance was set to 15 ppm, whereas fragment mass tolerance was set to 0.05 Da. The maximum false peptide discovery rate was specified as 0.01.
To identify protein carbonylation sites, we enriched Prx6 by immunoprecipitation from cultured human cells. The Prx6 immunoprecipitation samples were processed for digestion by trypsin and the tryptic peptides were analyzed by nanoLC-MS/MS analysis and protein sequence alignment to identify proline sites conversion into glutamic acid in Prx6. The conversion was identified based on a mass shift of + 31.990 Da at the proline residue (Figures 2A and B). The experiments led to the identification of one specific site at Pro 45 in human Prx6 protein (Figure 2C).
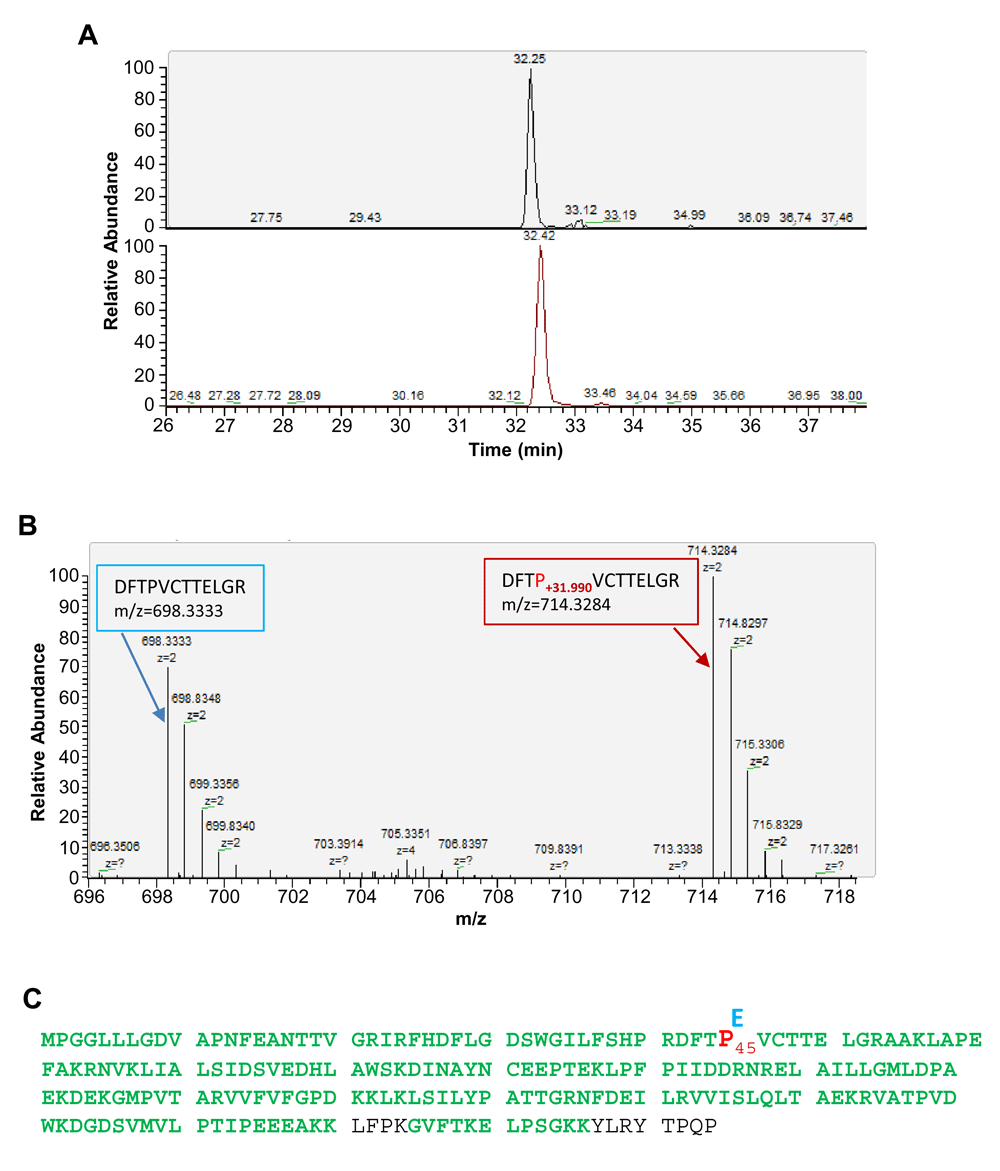
(A) Extracted ion chromatograms of Prx6 peptide (DFTP+31.990VCTTELGR, +2 charge, m/z=714.33) (top) and its non-conversion counterpart (DFTPVCTTELGR, +2 charge, m/z=698.33) (bottom). Both peptides were eluted at the same retention time and are from affinity-enriched cultured human cell extract using the anti-Prx6 antibody. (B) High resolution MS spectra of the co-elution of peptides (DFTP+31.990VCTTELGR, +2 charge, m/z=714.33) (right) and its non-conversion counterpart (DFTPVCTTELGR, +2 charge, m/z=698.33) (left). (C) Illustration of the identified proline 45 conversion into glutamic acid in cultured human cells (shown in bold red). Sequence areas containing amino acid residues shown in green are detected by LC-MS/MS analysis after trypsin digestion.
We are reasonably confident that the identified mass shift of + 31.990 Da is caused by the conversion of proline into glutamic acid, since the Prx6 was affinity-purified before MS/MS analysis. Since the conversion of proline into glutamic acid in Prx6 is a novel post-translational modification identified so far, it is desirable to confirm the structure of the identified peptides to ensure that the derived mass shifts of +31.99 Da are caused by the conversion into glutamic acid. MS/MS and HPLC co-elution are gold standards for verifying peptide identification. As demonstrated in Figure 3, both peptides, DFTP+31.990VCTTELGR, +2 charge, m/z=714.33, and its non-conversion counterpart DFTPVCTTELGR, +2 charge, m/z=698.33 were co-eluted with a peak shift of less than 0.2 min. Our result showed that the high resolution MS/MS fragmentation patterns of DFTP+31.990VCTTELGR and its non-conversion counterpart DFTPVCTTELGR peptide were almost identical except the addition of +31.990 Da of fragments that contain the proline 45 residue (Figures 3A and B).
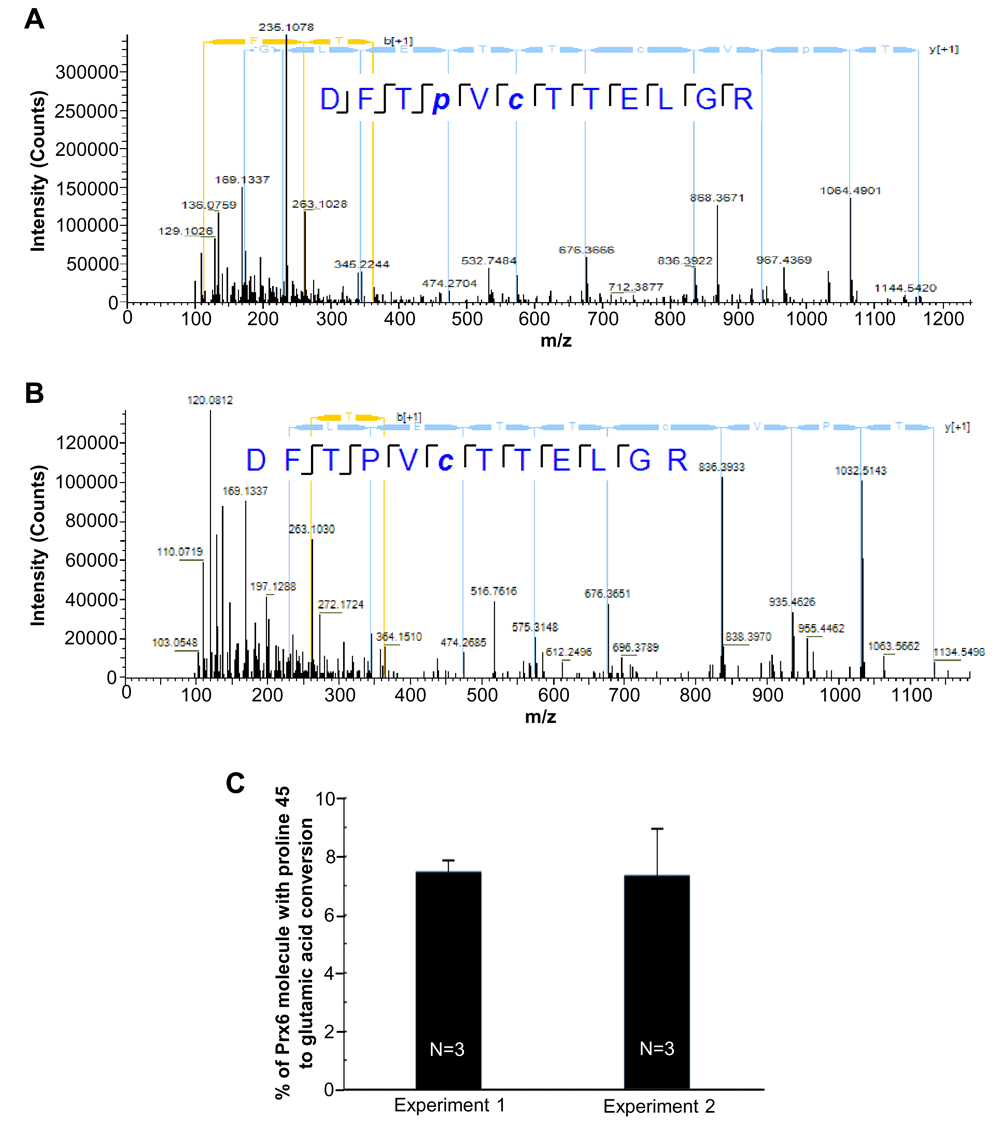
(A) High resolution MS/MS spectra of peroxiredoxin 6 (Prx6) proline to glutamic acid conversion peptide (DFTP+31.990VCTTELGR). (B) High resolution MS/MS spectra of Prx6 proline 45 peptide (DFTPVCTTELGR). Spectrum was obtained by LC-MS/MS analysis using the Thermo UltiMate 3000 RSLCnano System and Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer. (C) % of Prx6 molecules with the proline 45 conversion into glutamic acid in cultured human cells. Two independent MS analyses in triplicate (a total of six cell samples) were performed.
Analysis of the ion intensity of the MS spectra of DFTP+31.990VCTTELGR and its non-conversion counterpart DFTPVCTTELGR peptide (Figure 3C) determined that the proline 45 to glutamic acid conversion occurs in 5–10% of the Prx6 molecule in our samples with a mean of 7.43 ± 1.78% (N=6).
The present study introduces a revolutionizing concept that a protein engineering-like process could occur naturally in the biological system. Specifically, we identified that proline 45 of the Prx6 protein can be converted into glutamic acid. Proline 45 is in the peroxidase catalytic domain (Fisher, 2011; Fisher, 2017), thus this conversion should have functional significance. Future work should identify if this conversion increases, decreases or modifies the catalytic activity of Prx6. Such studies would open up the possibility that proteins with altered amino acid sequences have functional roles in the biological system.
The results from the present study also open up a new mechanism of ROS, indicating that the amino acid conversion, specifically the proline–glutamic acid conversion, is a consequence of oxidative stress mediated by the formation of glutamyl semialdehyde in the process of protein carbonylation. Through glutamyl semialdehyde, other conversions among arginine, proline, and glutamic acid are possible. Since the caged and site-directed production of hydroxyl radicals and carbonyl formation can occur via metal binding to specific sites of the protein structure (Stadtman & Berlett, 1991; Wong et al., 2010), ROS-mediated amino acid conversion may be a tightly regulated process.
The raw MS files from the output of the LC/MS/MS are available: doi, 10.17605/OSF.IO/5FN2E and 10.17605/OSF.IO/RP9J8 (Suzuki, 2017a; Suzuki, 2017b).
YJS conceived the study and designed the experiments. JH and YJS carried out the research. JH and YJS prepared the first draft of the manuscript. Both authors were involved in the revision of the draft manuscript and have agreed on the final content.
This work was supported by the National Institute on Aging and National Heart, Lung, and Blood Institute (NIH; grants R03 AG047824 and R01 HL72844, respectively) to YJS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 28 Sep 18 |
read | ||
|
Version 1 28 Apr 17 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)