Keywords
Migration, Quantification, User-Friendly, Microscopy.
Migration, Quantification, User-Friendly, Microscopy.
In order to understand and treat cancer, we need to study and ultimately control metastasis1. A key aspect of metastasis is cell migration1. Thus, assays that can reliably provide quantitative readouts of cell migration are an important component of cancer research. Here we describe an integrated computational pipeline to quantify cell migration using fluorescent microscopy.
The anterior gradient protein 2 (AGR2) has been shown to promote cell migration2,3. High expression of AGR2 is correlated with aggressive forms of various adenocarcinomas including prostate3 and breast cancer4,5. Therefore, AGR2 is a biomarker and therapeutic target that will provide a suitable biological readout of cell migration for our pipeline to quantify. In other words, because AGR2 is known to promote cell migration, it is ideal for testing a computational platform’s ability to detect changes in cell migration.
In this work, we describe an experimental and computational pipeline to quantitate cell migration. We demonstrate this pipeline by quantifying migration of MDA-MB-231 cells, a breast cancer cell line known to migrate aggressively6. We show that blocking the extracellular AGR2 (eAGR2) with a neutralizing antibody that binds specifically to AGR2 (referred to as AGR2-Ab) in cell medium prevents the migration of the MDA-MB-231 cells3. Our pipeline aids in the verification of well-established hypotheses and it can be used to test new hypotheses, thus aiding in and accelerating the drug discovery process.
MDA-MB-231 cell line was obtained from the American Type Culture Collection (ATCC, No. HTB-26) and tested for mycoplasma contamination before usage. Cells were maintained in DMEM media (Thermo Fisher, No. 21063045), supplemented with 10% fetal bovine serum (FBS), they were kept at 37°C in a humidified incubator with 5% CO2. Cells were used within 6 months of purchase.
The Oris™ migration assay (No. CMA1.101, Platypus Technologies, Madison, WI, USA) uses a physical barrier “stopper” to create a defined circular region that is intended to prevent cell adhesion at the start of the assay. This central cell-free detection zone is in the center of each well of a 96-well plate. As the cells migrate to the cell-free zone over 24–48 hours, real-time assessment of migratory cells allows acquisition of richer data sets. Since there are no artificial membranes or inserts in the light path through which cells must pass, this assay is amenable to quantification with microscopy. We used the Oris cell migration assay from Platypus Technologies to create migration regions by inserting stoppers in each of the 96 wells on a plate. Shortly after inserting the stoppers, we seeded MDA-MB-231 cells and waited until they reached 80% confluent (approximately 24 hours). We then fed cells with either treated or untreated media. Treated media included Taxol (5 nM), mouse anti-AGR2 antibody (1:50 of 2 μg/ml) (Santa Cruz Biotechnology, No. sc-101211), and mouse-IgG (Santa Cruz Biotechnology, No. sc-2005), while the untreated media was Dimethyl sulfoxide (DMSO) (VWR, No. 97061-250). Next we removed the stoppers and allowed the cells to move into the migration region. 48 hours after removal of the stoppers, we imaged each well using a fluorescence microscope (Zeoiss Observer.Z1 microscope at 2.5x objective with AxioCam MRm camera and Axio Vision 4.8 software).
This tool reads microscopy images which are placed in the same folder as the main code. First, the script first trains using the negative control (i.e., the image of a well where the stopper was not removed) to find the optimal disk which represents the migration region. Next, the script finds all the images present in the same folder as the code (by default it looks for TIF files) and applies the migration quantification metric to each of them, recording its output in a text file called output_c3.txt for easy access and plotting (a Python 3 script which creates a publication-quality plot based on this output is provided as well for any user who wishes to use it).
Typically a user only needs to make one modification the main script called code.m, make one minor modification to the img_name variable (on the third line of the script), and run the script. The change to the img_name variable needs to reflect the name of the image which contains the negative control.
If the user desires change the format of the images the script uses for the quantification, the line file_list= dir('* .tif'); needs to change to reflect the desired format. The images need to be in the same directory as the script.
This tool has been tested in multiple laptops running Matlab spanning releases 2015a–2018a.
We first created a mask corresponding to the area covered by cells using standard deviation filtering and applying a series of morphological operations in Matlab R2016a (MathWorks) as shown in Figure 1.

In order to identify the migration region, we took images of each well (left), then we select a mask that covers the area utilized by cells, highlighted in green (right).
Note that these images are gray scale (green is used throughout to highlight software outputs as is shown in the right panel of Figure 1), hence every pixel’s value belongs to the interval [0,1] where a completely black pixel has value 0 and a completely white pixel has value 1. Also note that a mask is a binary matrix that indicate which pixels belong to the mask (with value of 1, these pixels are referred to as “cell pixels”) and which pixels do not belong to the mask (with value 0).
We then used a genetic algorithm to determine the coordinates of the center and the radius of a circle according to Equation (1). This optimal circle determines the migration region.
where and r* are the optimal parameters of the migration region, M is the Matlab mask we are evaluating (i.e., a circle with center at coordinates (cx, cy) and radius r), so is the sum of all the pixel intensities (mi,j) which belong to the mask M, #M is the cardinality of M (i.e., the number of pixels which belong to M), and p is a penalty parameter. If p = 1, we have:
Hence, the maximization problem from Equation (1) is equivalent to the minimization represented in Equation (2) (when p = 1). From Equation (2), we can interpret the optimization performed by the genetic algorithm as finding “the largest circle which contains the least number of cell pixels.” Figure 2 shows the optimal circular region selected by the genetic algorithm when the input is the image from Figure 1.

The genetic algorithm selects the largest circle which contains the least number of masked pixels from the cell area mask. This optimal circle is the migration region.
Percent of migration region covered by cells: We defined a metric to quantify the migration of MDA-MB-231 cells. This metric is Q, the percentage of migration pixels inside the migration region. We define a migration pixel as any pixel whose intensity value is greater than or equal to a threshold T. We chose T equal to 1.25 times the median pixel intensity of the migration region immediately after the stopper was removed (i.e., the green region in Figure 2). This is:
Where M* is the optimal circle defined by the three parameters ( from Equation (1) and Equation (2)) and the set {M* ≥ T} includes all of the pixels inside M* with intensities higher than T .
A previous version of this manuscript is available from bioRxiv: https://doi.org/10.1101/1305267
To test the hypothesis that MDA-MB-231 cells’ migration is reduced in the absence of AGR2, we designed an experiment (utilizing the cell migration assay described in the Methods section) with 5 experimental conditions: a positive control (untreated cells), a negative control (wells where the stopper was not removed), cells treated with 10nM of Taxol (a non-cytotoxic dose level which prevents cell migration but does not promote cell death8), cells treated with a 1:50 dilution of AGR2-ab to inactivate eAGR2, and with a 1:50 dilution of IgG, a control antibody (Ctrl-Ab) which does not affect cell migration. Representative images from these conditions (i.e., replicate 1) are shown in Figure 3 (top).
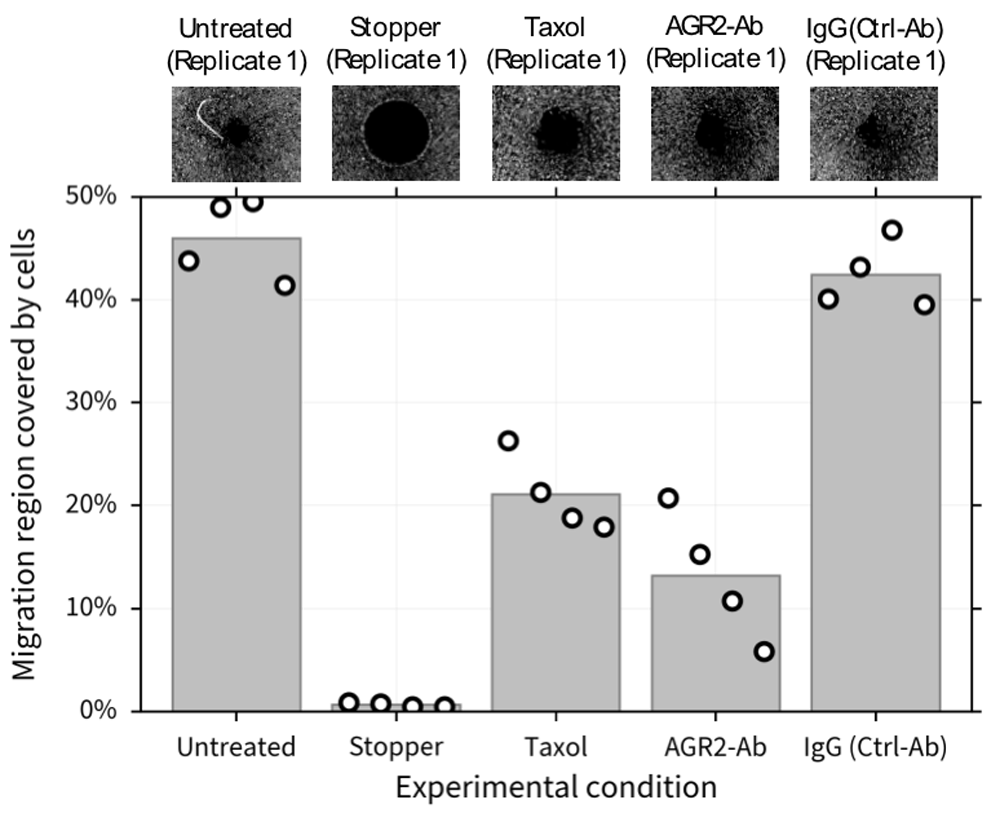
Representative images (replicate 1) of each condition are shown (top). 10nM of Taxol and the 10µg/mL of the H10 peptide show similar levels of migration inhibition compared to the positive and negative controls. Our metric (bottom) allows us to quantify the qualitative results (top).
For the untreated case and the control peptide we observe increased migration, with 46±2 (mean ± standard error of the mean) percent of the migration region covered in the untreated case and 42±1.7 percent of the migration region covered in the control antibody case. We fail to reject the null hypothesis that these two are the sample means from the same distribution (p value of 0.084). In the Taxol case, 21±1.9 percent of the migration region is covered. We reject the null hypothesis that the mean of the Taxol population and the mean of the untreated case are sample means from the same distribution (p value of 8.16e-5). Similarly, for the AGR2-Ab case, 13±3.2 percent of the migration region is covered. We reject the null hypothesis that the mean of the AGR2-Ab population and the mean of the untreated case are sample means from the same distribution (p value of 2.19e-5). Not only do we confirm the hypothesis that MDA-MB-231 cells’ migration is reduced in the absence of AGR2, but our method allows for reproducible quantification of these qualitative observations. Furthermore, the algorithms used to compute Q requires a single input from the user (a string with the names of the control experiments) and it can run on a desktop machine with Matlab (R2015a-2018a and above) installed.
We have designed and implemented a pipeline for quantifying cell migration in vitro. It is worth noting that this metric may not discern between cell motility and proliferation, hence in order to use it to estimate parameters for a mechanistic model, a parameter estimator such as CellPD9 should be used to estimate parameters of processes which decouple proliferation and motility10,11. However this metric is robust to image noise, open source, replicable, replicable and user friendly. In particular, we show that H10 aids in the reduction of migration of MDA-MB-231 cells by blocking sAGR2. This pipeline can be expanded to various cancer cell lines and model systems.
Raw, unedited microscope images from this analysis are available from the project GitHub repository: https://github.com/edjuaro/cell-migration-quantification
The source code used throughout this manuscript can be accessed in the public GitHub repository: https://github.com/edjuaro/cell-migration-quantification.
Archived source code at time of publication can be found here: http://doi.org/10.5281/zenodo.132392312
This code is released under the permissive MIT license.
This work was supported by the USC Center for Applied Molecular Medicine, the National Institutes of Health (Physical Sciences Oncology Center [5U54CA143907] for Multi-scale Complex Systems Transdisciplinary Analysis of Response to Therapy (MCSTART), and [1R01CA180149]), the Breast Cancer Research Foundation, the USC James H. Zumberge Research and Innovation Fund, and a USC Provost’s PhD fellowship.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Partly
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
References
1. Gebäck T, Schulz MM, Koumoutsakos P, Detmar M: TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays.Biotechniques. 2009; 46 (4): 265-74 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Computational cell dynamics Quantitative cell biology Cell migration High throughput phenotyping Computer vision application
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
No
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 15 Aug 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)