Keywords
database identifier mapping, gene variant, BridgeDb, interoperability
This article is included in the ELIXIR gateway.
database identifier mapping, gene variant, BridgeDb, interoperability
Many bioinformatics software tools rely on database identifier mapping, for instance for 1) recognition and mapping of identifiers used in experimental data to the corresponding identifiers present in secondary sources like pathways or ontology classes or 2) simply to combine data from different sources that use different identifiers. BridgeDb is a database identifier mapping tool that is available as a Java framework and as an installable web service (van Iersel et al., 2010). Tools that integrate BridgeDb are for instance: the community curated pathway resource WikiPathways (Slenter et al., 2018), the modular pathway editor and pathway analysis tool PathVisio (Kutmon et al., 2015), and the network tool Cytoscape used to visualize, extend and evaluate biological networks. Depending on the available mappings BridgeDb can provide the mapping between identifiers from various data sources, also when these link to different molecular levels, e.g. gene to protein. BridgeDb can also be deployed as a web service. Moreover, it is available in a semantic web version, the Identifier Mapping Service (IMS), which can be used inside the Open PHACTS platform but can also be deployed from a software container (Gray et al., 2014) (link to tutorial and link to GitHub). Mappings for BridgeDb are already available for gene products for many species (produced from the respective Ensembl genome annotations (Aken et al., 2017)), for metabolite identifiers (produced from HMDB (Wishart et al., 2013)) and ChEBI (Hastings et al., 2013)), and for reaction identifiers (produced from Rhea (Morgat et al., 2017)).
The BridgeDb mapping databases are linking pins between tools that support genetic variants, genes, and pathways analysis helping to visualize a complex biological context such those typical of the multifaceted (genetic) diseases. Gene-to-variant mapping was not yet available for BridgeDb. Such mappings can be especially useful to work with genetic variations, for instance when evaluating traits with a complicated genetic background like blood pressure, susceptibility to heart failure, or diabetes type 2 development. Single nucleotide polymorphism (SNP) can be responsible for phenotypic variations. In extreme cases this can be the cause of rare genetic disorders. For example, several SNPs in the human DMD gene can cause Duchenne muscular dystrophy (DMD), a severe congenital disorder which leads to severe physical impairment (Magri et al., 2011). Since BridgeDb can stack mappings, the combination of the new gene-to-variant mapping database with the collection that was already available offers versatile mappings for variants to a large set of different human gene and gene product identifiers.
The main objective of this work was to provide mappings between gene identifiers and variant identifiers in both directions. The steps needed to achieve this were: 1) select the best source for the mappings, 2) collect data from the selected source, 3) annotate the result with provenance data about the process, the source, and the source version, and 4) finally to release the new BridgeDb mapping database and integrate that in the regular BridgeDb mapping database update schedule.
Target users for the resulting mappings are 1) bioinformaticians and developers, working on new approaches for data integration, if these use human genetic (variant) information; 2) members and users of ELIXIR data interoperability services, including the implementations in the tools mentioned that perform analyses based on human genetic variant data, for instance for the analysis of common multifaceted genetic diseases or in the rare disease field; and 3) researchers who access and query molecular data resulting from the analysis above.
The gene-to-variant database uses mappings between Ensembl and dbSNP (Kitts et al., 2013). The Ensembl gene-to-dbSNP variant mappings present in Ensembl were used as the source. The released database is based on Ensembl r91, dbSNP b150, and the human genome assembly GRCh38. Although Ensembl provides more genetic variation from different sources, we focused on dbSNP as this variation database is regularly updated and adjusted to the actual Ensembl genome built. We compared both sources (Ensembl and dbSNP) and made sure that Ensembl provides all dbSNP available variants. So, we are able to rely only on the Ensembl API as a source for the extraction of the data necessary for creation of this mapping database. To prevent problems introduced by the user interfaces we used database dumps for this comparison.
The data dump was obtained from the Ensembl ftp server (link for download). For the first version, we used Ensembl 91, gene annotation with Gencode 27. The vcf (variant call format) file is the one relevant for our mapping. It contains the dbSNP identifier with its additional attributes and the associated Ensembl transcript identifier. By querying the Ensembl platform web service, we can access the gene identifier of the transcript. Combined, this leads to mappings between variants and genes. The size of the complete mapping database exceeded 150 Gb (for Ensembl 91), so we decided to create several different subsets: exonic variants, missense variants, protein truncating variants (PTV), PTV and missense variants, and variants with a PolyPhen score >0.908 indicating “Probably Damaging”. Other selections can be created easily on individual demand.
The created database contains the link between the Ensembl gene identifiers and the dbSNP variant identifiers including a selection of attributes (MAF (minor allele frequency), chromosome, variant alleles, and chromosome position start/end).
For the rare cases where a variation is associated to more than one gene, the variant is also associated to these genes in the BridgeDb database. For example, rs199773918 overlaps in the exons of two genes (ENSG00000173366 and ENSG00000239732), and in the exonic variant BridgeDb mapping both genes show up. Nevertheless, in our selection of variants it may happen that not all of them show up due to different variant effect classifications in the different genes. As an example, rs199773918 is a variant that overlaps in the following genes: TPR (ENSG00000047410) and PRG4 (ENSG00000116690). This variant is a “3’ prime UTR variant” of TPR and a “missense variant” of PRG4. It can be found in both genes variant tables but due to our selection it will show up only once in the missense variant dataset.
Database creation. An open-source Java program to create the gene-to-variant database is available on GitHub. After downloading the vcf file from Ensembl, users create a configuration file with several parameters. Then the database creation program will parse the vcf file, retrieve additional information through the Ensembl web service and create the BridgeDb mapping database. Due to the large amount of mappings, the tool commits the mappings to the database in batches to keep the required memory low.
Operation. The database creation workflow is depicted in Figure 1. The vcf file can be downloaded from the Ensembl FTP. The “Homo_sapiens_incl_consequences.vcf.gz” file is used.
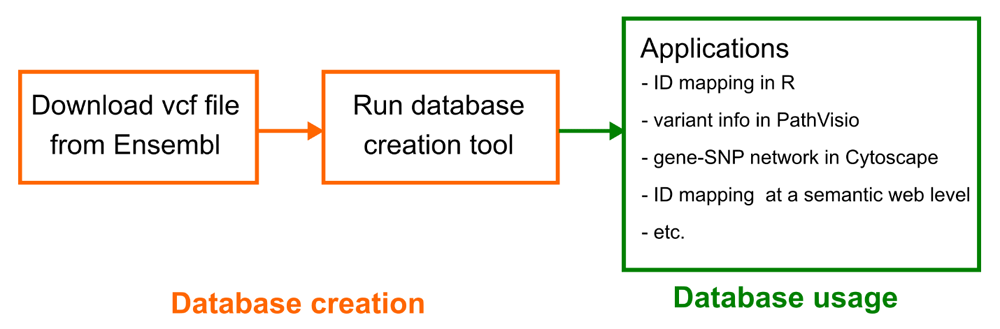
The gene-variant mapping database is built on the variant call format (vcf) file provided by Ensembl. After running the database creation tool, the database can be used in all the different use cases.
System requirements. The database creation tool runs with Java and requires more memory than usually given to a Java process. We advise users to allocate 3–4GB of memory at least when running the database creation tool (-Xmx4G).
The resulting BridgeDb mapping databases are available as a Derby database from here. The new mappings are available for all the BridgeDb implementations mentioned above (PathVisio, Cytoscape, R package, web service, and the IMS). The mapping databases are freely available for download under CC-BY license. Application examples of the use of the variant BridgeDb database are given in the following section. We created gene-to-variant mapping databases for the variant classes given in Table 1. Any other subset of variant classes can be created on demand using the tool described in the Methods section.
| SNP selection | File | Date | Size (zipped) |
|---|---|---|---|
| Exonic variants | SNP_r91_Exon.bridge.zip | 2018-06-04 | 1.1Gb |
| Missense variants | SNP_r91_Missence.bridge.zip | 2018-06-07 | 620Mb |
| Protein truncating variants | SNP_r91_PTV.bridge.zip | 2018-06-07 | 75Mb |
| Protein truncating variants and missense: | SNP_r91_PTV_Missense.bridge.zip | 2018-06-07 | 620Mb |
| All variants with a PolyPhen score > 0.908 indicating “Probably Damaging” | SNP_r91_PolyPhen.bridge.zip | 2018-07-18 | 260Mb |
To test and demonstrate the application of the variant BridgeDb database, we downloaded the database from BridgeDb. The gene-to-variant (and variant-to-gene) queries are shown in four different tools: R command line (Team, 2014), PathVisio (Kutmon et al., 2015), Cytoscape (Shannon et al., 2003) and the local IMS installation using Docker, in order to provide an overview of the flexibility of the mapping database in different environments. A genetic variant of the rare disease Duchenne muscular dystrophy (DMD) was selected from the gene-disease association database DisGeNET (Piñero et al., 2017). The rs104894790 (Lenk et al., 1993) SNP was chosen because it presented a high number of citations and a stop gain damaging effect on the gene’s protein product.
The SNP, rs104894790, as described above was used to query the Ensembl identifier for the gene(s) in which it is located (variant-to-gene query). The query was performed in R command line, after the installation of the BridgeDb R package (link to BridgeDb R package) (example R script in Supplementary File 1) (R version 3.5.1). The result shows that the variant is positioned only in one gene: dystrophin (DMD, ENSG00000198947). DMD is one of the largest genes in the human DNA (about 2.2 Mb), and is composed of 79 exons and has 32 known transcripts of which 20 are protein coding. Because the output is identifiers, it can be easily linked to other R packages such as mygene (Mark et al., 2014) which normally wraps around the mygene.info web service (Xin et al., 2016).
We used PathVisio (version 3.3.0) (Figure 2), a biological pathway analysis tool that allows drawing, editing and analyzing biological pathways, to demonstrate how the new gene-variant database can be used to evaluate variants in a pathway context. PathVisio, like Cytoscape, has the BridgeDb functionality integrated in the core. For the purpose of the demonstration, we first selected pathways that contain the DMD gene from the R example. Five pathways were found: two striated muscle contraction pathways (WP3795 and WP383), Ectoderm differentiation (WP2858), Extracellular matrix organization (WP2703) and Arrhythmogenic right ventricular cardiomyopathy (WP2118). In principle, a new PathVisio plugin could now be developed that searches pathways that contain genes with selected variants automatically, or the plugin could show all variants from an analysis sets on a given pathway. For the example, one of the striated muscle contraction pathways (WP383) was selected and visualized. Next, the BridgeDb variant database was loaded, using the BridgeDbConfig plugin. After selecting a gene in the pathway, the backpage tab of the right hand side panel now shows the list of hyperlinks obtained from the BridgeDb database that point to different information sources linked to the gene selected. Figure 2 shows the backpage with the list of the 720 SNPs (from the BridgeDb with a PolyPhen score > 0.908, file SNP_r91_PolyPhen.bridge) for the selected DMD gene. All the SNPs in the backpage have a hyperlink to the corresponding dbSNP page.
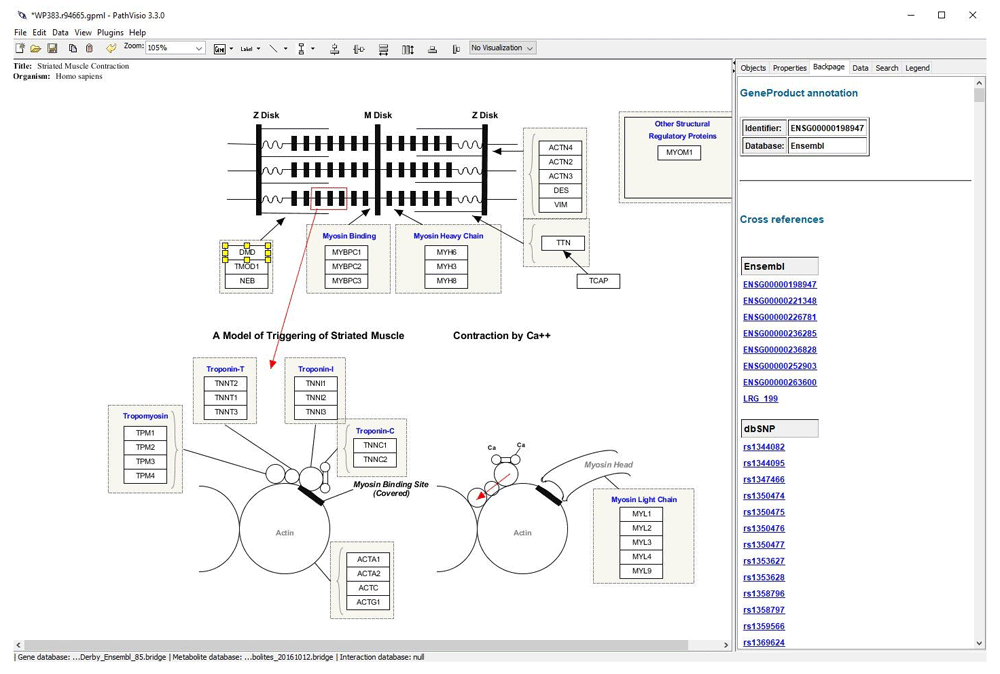
When the DMD gene is selected a list of hyperlinks from different sources are displayed in the back page of the left panel. In this case, a list of SNPs located in the gene is visualized.
An alternative gene-to-variant visualization is provided using Cytoscape (version 3.6.1), a popular tool for (biological) network analysis and visualization (Figure 3). The BridgeDb app for Cytoscape is available here. A node with the Ensembl gene identifier of DMD was created and the 720 SNPs were mapped to the gene using the BridgeDb app interface. A gene-variant network was created using the list of variants mapped. Moreover, the app can be used to configure the selection of several attribute columns related to the variant nodes such as: chromosome location, minor allele frequency, and variant allele. In this example figure, we visualize the PolyPhen score as the node fill color of the variants. For simplicity, the rs-numbers are not displayed.
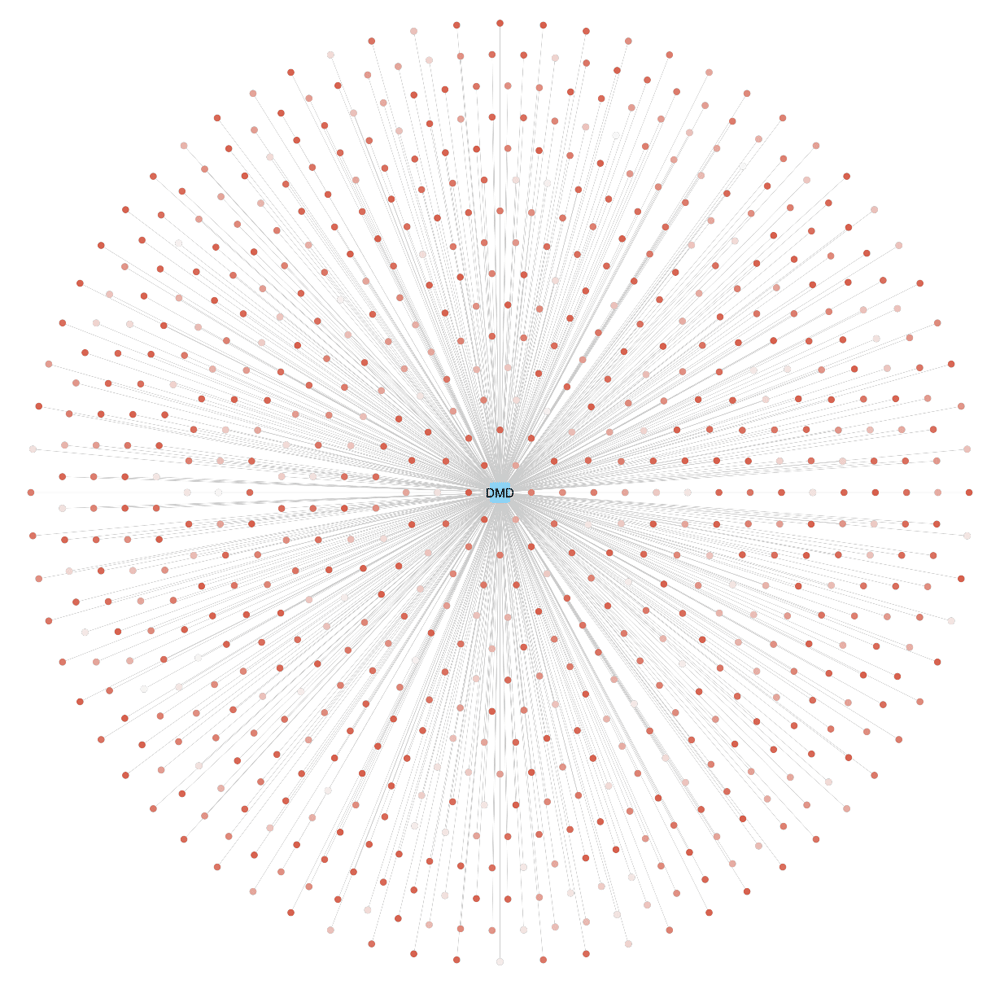
Using the BridgeDb app for Cytoscape, a gene-variant network for the DMD gene (blue rounded rectangle) and its 720 probably damaging variants (PolyPhen score > 0.908) was created. The node color of the variants represents the PolyPhen score as a gradient (white-red), the darker the red, the higher the PolyPhen score.
Finally, we here show that identifier mapping linking variants to genes and vice versa can also be done at a semantic web level, we here demonstrate how an online BridgeDb Identifier Mapping Service (IMS) can be set up. The IMS technology was developed in the Open PHACTS project to link drug discovery related data sets, including a Docker image (Batchelor et al., 2014; van Iersel et al., 2010; Williams et al., 2012). Here, identifier mappings are defined by link sets, which specify which identifiers are mapped. However, unlike traditional BridgeDb mapping files, these link sets also specify why the two identifiers are mapped, allowing them to be used as scientific lenses (Batchelor et al., 2014).
Because the IMS works at a semantic web level, identifiers are represented by uniform resource identifiers (URIs). Moreover, the IMS is aware of URI equivalence defined by the MIRIAM registry (Juty et al., 2012). This means that even when a mapping file does not provide mappings for a certain URI, one would still get a number of equivalent URIs, following knowledge from MIRIAM database. And, when a single mapping is found in the link sets, equivalent URIs for the mapped URIs it returned. The IMS provide a targetUriPattern parameter allowing you to restrict the number of mapped URIs.
We developed a tutorial explaining how to set up an IMS instance with the variant-gene mappings (available from GitHub). The instance is run locally using a Docker container developed by Open PHACTS, which is available from DockerHub. After the Docker image is started, it provides a web interface and an API. The web interface has a "Check Mapping for an URI" page where the URI can be given to be mapped, the return format (XML, JSON, or HTML), and optionally a lensUri (see (Batchelor et al., 2014), and the aforementioned targetUriPattern).
However, it is more convenient to use this API from other tools, as demonstrated with a second R script (Supplementary File 1). This R script uses the curl (Ooms, 2017 link) and jsonlite (Ooms, 2014) packages to interact with the IMS. The first package is used to call the IMS webservice and the second to convert the returned JSON into a data model more easily handled in R. The example consists of two API calls: the first part finds 603 variants for the DMD gene (Ensembl ID ENSG00000198947); the second example takes a single variant (dbSNP ID rs769658853) and looks up the matching gene.
The BridgeDb toolset provides several apps and tools designed for different purposes, while mapping databases are available to link different database IDs for genes and gene products, metabolites, and reactions and interactions. A mapping database in the BridgeDb software environment, capable of linking genes to their variants and vice versa, was not yet available. The new database is expected to be useful to enhance the biological interpretation of genetic variant data (as shown with the example of the DMD gene) for instance when using apps that evaluate biological pathways, use the classification of genes according to ontology terms, or in the R environment when performing gene and variant related statistical evaluation.
With this newly created mapping database and the transitivity function of BridgeDb, the user can map between three different layers: e.g. variant-gene-protein. This approach can support multi-omics analysis for various biomedical applications, and tools like Cytoscape and PathVisio can be used immediately to benefit from this.
We intend to keep the content up-to-date by regular updates. The human variant mapping database is already incorporated into the quarterly BridgeDb mapping database update. Also other variant sets including more than only the currently included protein truncating and missense variants can be created on user community (or individual) demand.
The new gene-to-variant mapping databases are available here: http://bridgedb.org/data/gene_database/
Available under a Apache 2.0 licence (http://www.apache.org/licenses/LICENSE-2.0.html)
Source code for making of the mapping databases is available from GitHub: https://github.com/BiGCAT-UM/BridgeDbVariantDatabase
Archived source code at time of publication: http://doi.org/10.5281/zenodo.1326514 (Willighagen & Melius, 2018)
License: Apache 2.0
This work was funded by ELIXIR, the European research infrastructure for life-science data.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would like to thank the BridgeDb development team. This work heavily leaned on previous work done by the dbNP and Ensembl teams who curated the actual mappings and on the original BridgeDb development team, especially Martijn van Iersel.
Supplementary File 1 – R code and instructions for setting up the BridgeDb IMS docker.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: bioinformatics
Is the rationale for developing the new software tool clearly explained?
Partly
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
No
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: bioinformatics, genomics, genetics, R
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 03 Sep 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)