Keywords
Bacteria, Thermoenzymes, Haloenzyme, Enzymes, Industrial Applications.
Bacteria, Thermoenzymes, Haloenzyme, Enzymes, Industrial Applications.
Covering large surface of the Earth's surface, the marine environment is a rich source of biological and chemical diversity; it contains endless habitats that may present adverse conditions of survival. However, these conditions favour the establishment of microorganisms able to produce enzymes that have extraordinary properties, such as salt tolerance, thermostability, pH and temperature variations. These enzymes have many industrial applications, such as the production of detergents, food, feed, pharmaceuticals, leather and biofuel1,2.
The conditions of the industrial scale activities are related to the maintenance of enzymatic activity in environments with variations in temperature (55°C to 121°C and -2°C to 20°C), pressure (> 500 atmospheres), pH (pH> 8, pH <4) and salinity (1–5 M NaCl or KCl)3. The production of enzymes of bacterial origin is a frequent application of industrial biotechnology; the enzymes produced include hydrolytic thermostable enzymes such as amylases, cellulases, proteases and xylanases for the production of biofuel4. Use of the genus Bacillus is promising for the production of biomolecules, because it is classified by the FDA as being generally recognized as safe and research has revealed the ability of this genus to produce and secrete enzymes with infinite applications5.
This study aimed to produce multiple thermoenzymes and haloenzymes (protease, cellulase, amylase and xylanase) expressed by Bacillus subtilis strain SR60, a bacterial symbiont isolated from Siderastrea stellate (Verrill, 1868) in a Brazilian coral reefs ecosystem 7°08’50” S; 34° 47’51” W.
The bacterial strains were obtained from aseptically collected tissues of Siderastrea stellate Verrill, 1868 (Cnidaria, Scleractinia) colonies at Cabo Branco coral reefs, Paraiba State, Brazil (7°08’50” S; 34°47’51” W). For bacterial isolation from the anthozoan, samples were suspended in sterile saline solution, agitated until homogenization was achieved and then spread over marine agar plates (pH 8.0± 0.3) containing 5 g/l peptone; 1 g/l yeast extract; 15 g/l agar diluted in sterile marine water and incubated at 55°C until adequate growth was achieved6. A total of 12 bacterial isolates were obtained, which were analysed for protease, cellulase, amylase and xylanase production capacity, and only the one with the simultaneous production capacity of these enzymes was selected.
For further screening of enzymatic activity described below, two bacterial colonies, isolated using the above culturing conditions, were inoculated onto each plate. A total of three replicates were performed for each salt molarity.
In order to identify the isolate, morphophysiological and molecular data were evaluated7. The obtained 16S rRNA gene was sequenced by ATCGene (UFRGS, Porto Alegre, RS, Brazil) using the automated sequencer ABI-PRISM 3100 Genetic Analyzer. The SR60 isolate sequence was compared to sequences deposited in the Genbank database (NCBI). For the local alignment, the BLASTn tool (NCBI) was used. MEGA 6.0 software was used for monitoring multiple sequences and for construction of a dendrogram by the Neighbor-Joining method.
The isolated bacterial strains were screened production for protease on agar medium comprising 10 g/l gelatine and 20 g/l agar in increasing concentrations of NaCl (0, 0.25, 0.50, 1.0, 1.25 and 1.5 M) pH 8.0± 0.3. The inoculated plates were incubated at 48 h at 55°C and observed for the formation of zone of hydrolysis8.
The ability of isolate on produce cellulose was tested a plate containing 1 g/l carboxymethylcellulose (CMC); 0.5 g/l NaNO3; 1 g/l K2HPO4; 0.5 g/l MgSO4∙7H2O; 0.001 g/l FeSO4∙7H2O; 1 g/l yeast extract; 15 g/l agar) in increasing molarities NaCl (0, 0.25, 0.50, 1.0, 1.25 and 1.5 M) for 48 h at 55°C on pH 8.0±0.3 and then overlaid with 0.2 g/l potassium iodide for 5 min, bacterial colonies showing clear zones were considered to be cellulase producers9.
Amylolytic activity of culture was screened on starch nutrient agar plates containing: 10 g/l starch; 0.05 g/l NaNO3; 1 g/l K2HPO4; 0.5 g/l MgSO4∙7H2O; 0.001 g/l FeSO4∙7H2O; 1 g/l yeast extract; 15 g/l agar, in increasing molarities of NaCl (0, 0.25, 0.50, 1.0, 1.25 and 1.5 M). After incubation at 55°C pH 8.0±0.3 for 48 h, the zone of clearance was determined by flooding the plates with 0.2 g/l potassium iodide for 5 min10.
Xylanase activity was detected using a saline medium containing: (10 g/l xylan; 0.005 g/l NaNO3; 1 g/l K2HPO4; 0.5 g/l MgSO4∙7H2O; 0,001 g/l FeSO4∙7H2O; 1 g/l yeast extract; 15 g/l agar) in increasing molarities of NaCl (0, 0.25, 0.50, 1.0 and 1.5 M) on pH 8.0±0.3. After incubation at 55°C for 48 h, the plates were with 0.2 g/l potassium iodide for 5 min. The clear zones around colonies indicated qualitative xylanase activity11.
The SR60 isolate was revealed to be a Gram-positive spore-forming bacillus, facultative anaerobe, catalase-positive; it was negative for indole, H2S production and citrate utilization bacterium (Table 1). Those findings led us to consider the isolate belonging to the genus Bacillus which was posteriorly confirmed by the phylogenetic analysis which revealed that the SR60 strain formed a clade with Bacillus subtilis (Figure 1). The nucleotide sequence was deposited in GenBank under accession number MH698455.1.
| Parameter | Result |
|---|---|
| Gram staining | Positive |
| Morphology | Bacillus |
| Arrangement | Absent |
| Endospore | Positive |
| Catalase | Positive |
| Urease | Negative |
| Citrate Utilization | Negative |
| H2S Production | Negative |
| Indole Production | Negative |
In differential media for the production of different extracellular enzymes, it was observed that conditions of high salinity from 0 to 1.5 M NaCl, a SR60 strain showed proteolytic, cellulolytic, aminolytic and xylanolytic activity, these productions being observed by zones of enzymatic hydrolysis (Table 2). The halo detection for protease and cellulase was observed up to the maximum salinity, 1.5 M NaCl (Figure 2 and Figure 3). Cellulolytic enzymes comprise a group of glycosidic hydrolases, including endoglucanases, exoglucanases and beta-glycosidase. In general, the production of the enzyme group is mainly observed in fungi, actinomycetes and some other bacteria. The use of fungi to produce cellulases has been practiced in the food, textile, fuel and chemical industry, but the growth period for the microorganism does not match the high demand from the industries for production. In an attempt to solve this problem bacteria present rapid growth and high enzymatic production12. Bacterial isolates produced from different environments, such as bovine ruminants, soil and in isolation, were found to produce hydrolases12,13. Biofuel industries that use lignocellulose as the first raw material pre-treatment process for the release of cellulose, making it more accessible to the enzymatic action. During the processing of the cellulose, various compounds containing salts are used, the enzymatic catalysis being reduced or inhibited in this halophilic environment15. The extracellular production of amylase and xylanase reached an upper NaCl concentration limit of 1.0 M and 1.25 M NaCl, respectively (Figure 4 and Figure 5); however, as a bacterial cell growth molecule at the other salt concentrations.
| Zone Formation | ||||
|---|---|---|---|---|
| Molarity NaCl | Pro | Cel | Amy | Xyl |
| 0 M | + | + | + | + |
| 0.25 M | + | + | + | + |
| 0.50 M | + | + | + | + |
| 1.0 M | + | + | + | + |
| 1.25 M | + | + | - | + |
| 1.5 M | + | + | - | - |
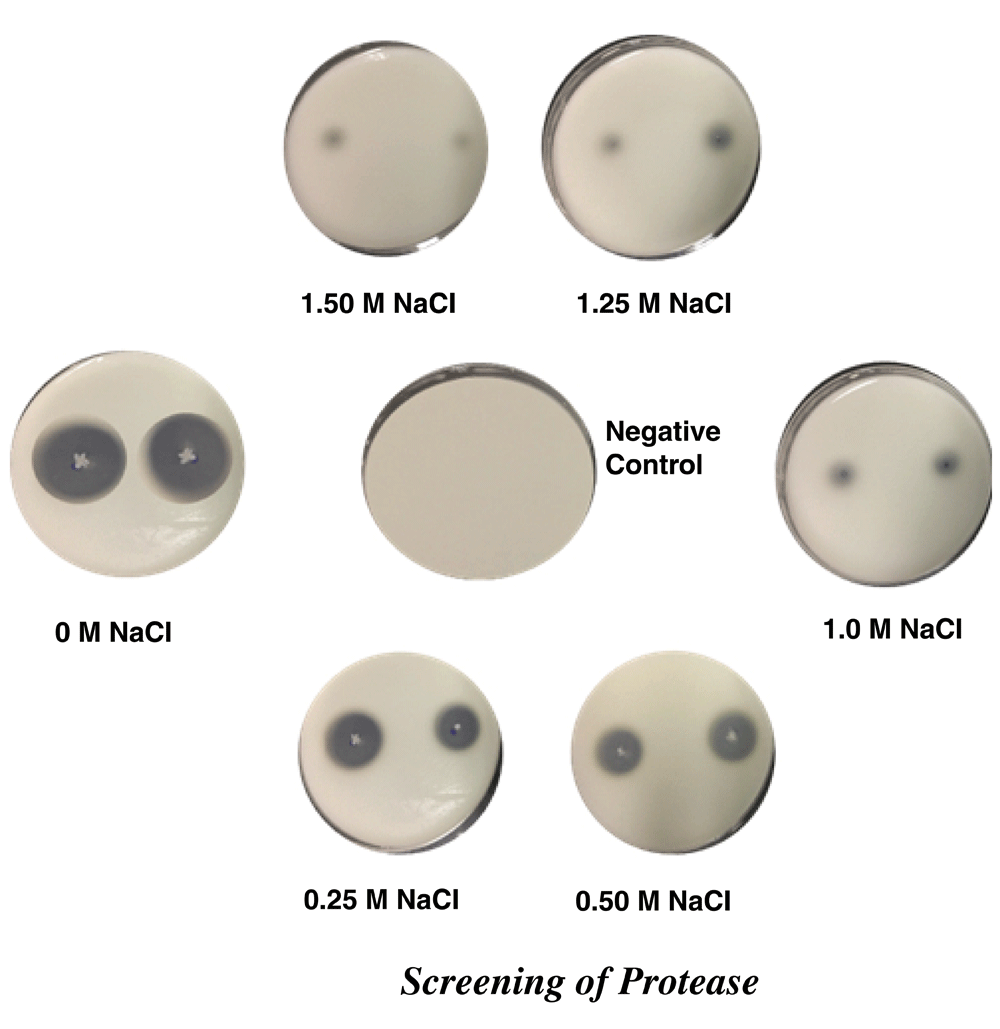
Halos around bacterial colonies are indicative of cellulose degradation.
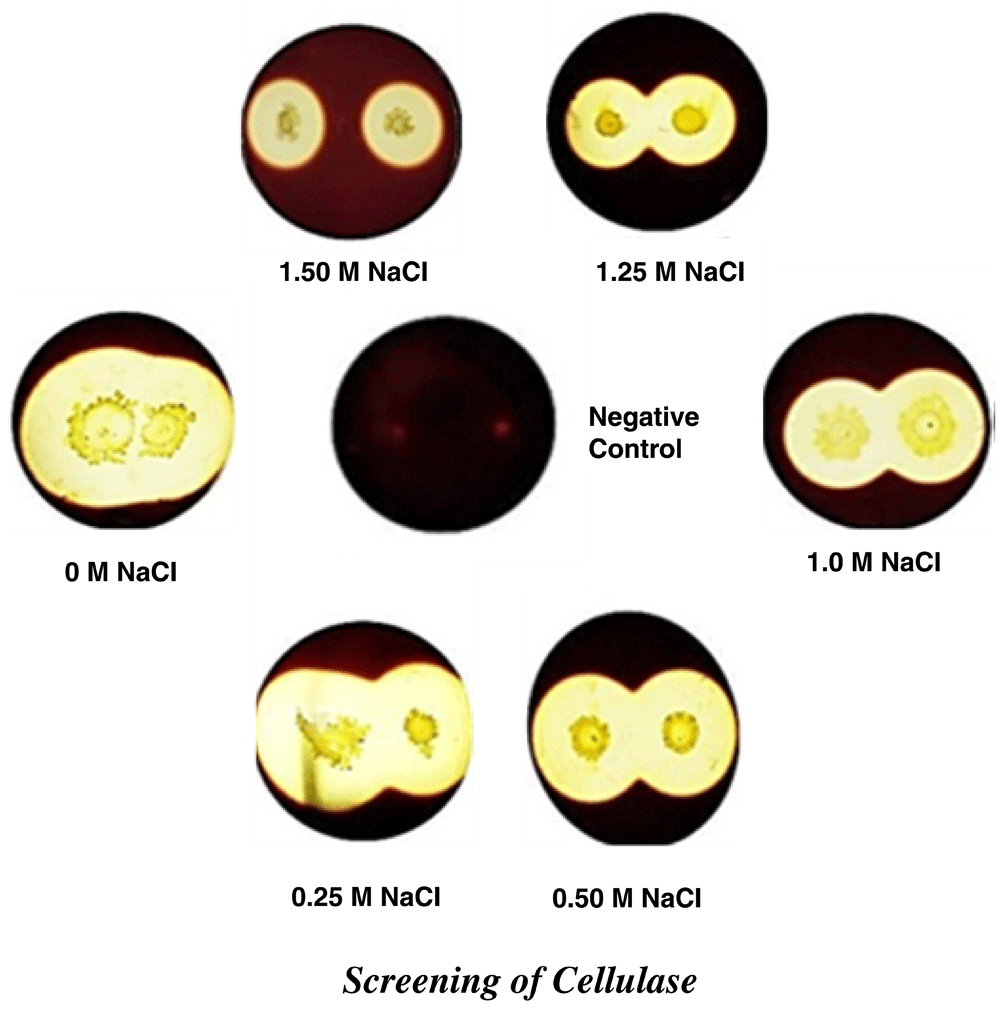
Halos around bacterial colonies are indicative of cellulose degradation.
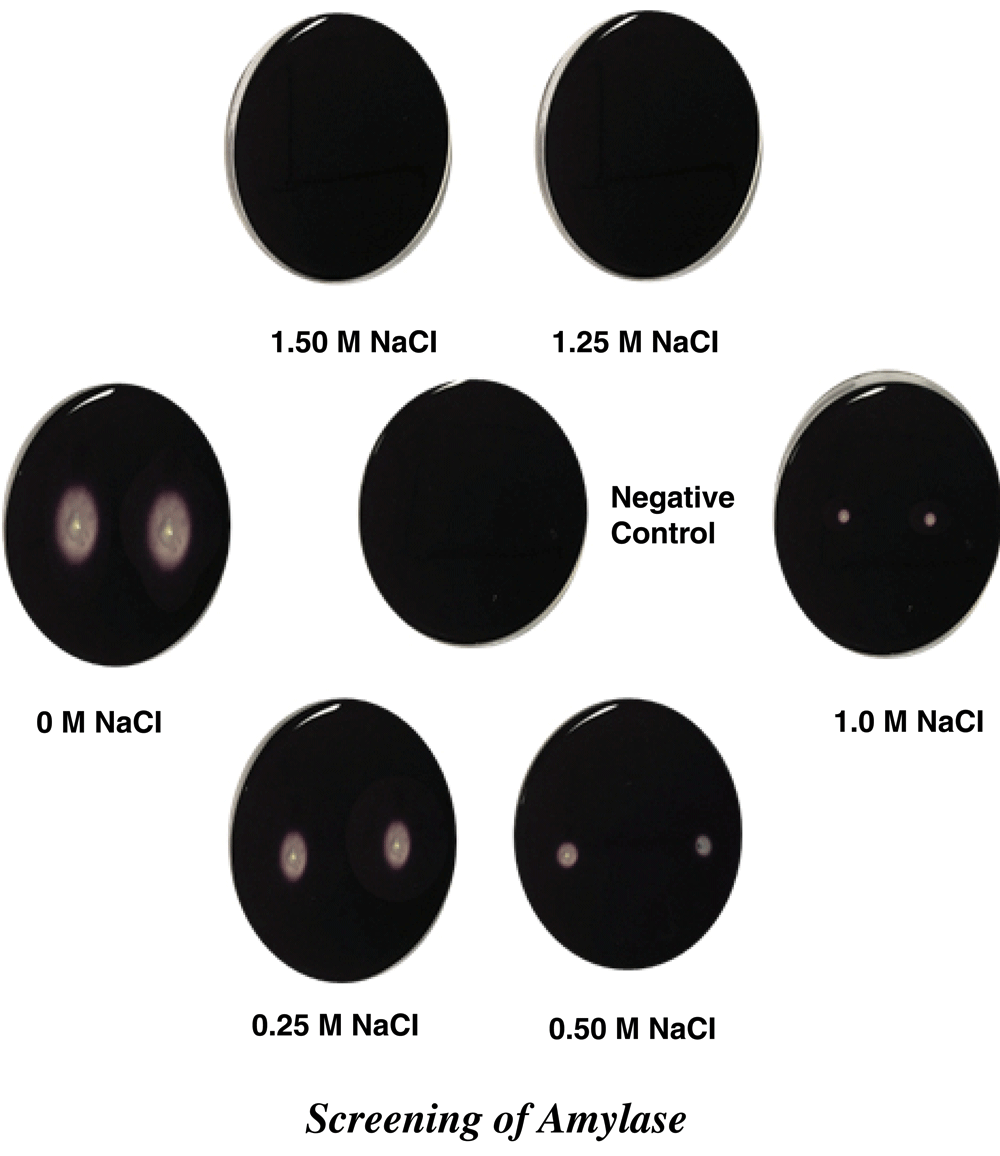
Halos around bacterial colonies are indicative of cellulose degradation.
The Bacillus sp. isolate identified in this study, Bacillus subtilis SR60, has the capacity for proteases, cellulases, amylases and xylanases with thermostable and halotolerant characteristics. These products can be used as thermostable enzymes in the production of biofuels in crucial stages of this bioprocess.
The sequence of the Bacillus subtilis strain SR60 16s RNA gene isolated in this experiment is available from GenBank, accession number MH698455.1: https://identifiers.org/ncbigi/GI:1435753077.
Images of the repeats of the screening for enzymatic activity have been uploaded to Harvard Dataverse, DOI: https://doi.org/10.7910/DVN/J5JCC0. Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication.
This work was supported in part by the Federal University of Pernambuco.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Ariaeenejad S, Sheykh Abdollahzadeh Mamaghani A, Maleki M, Kavousi K, et al.: A novel high performance in-silico screened metagenome-derived alkali-thermostable endo-β-1,4-glucanase for lignocellulosic biomass hydrolysis in the harsh conditions. BMC Biotechnology. 2020; 20 (1). Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Enzyme biochemical characterization
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 26 Oct 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)