Keywords
Yeast species, ITS sequencing, Cancer patients.
Yeast species, ITS sequencing, Cancer patients.
As a result of the immunocompromised state and the effect of chemotherapy, cancer patients are more susceptible to fungal infections, especially by Candida species (AI-Dwairi et al., 2014). Candida species are commensal in human bodies, and become opportunistic pathogens in immunodefective patients. Sometimes they can cause systemic infections and colonize different organs due to their dissemination from the mucosal infected regions (Miceli et al., 2011). Pfaller et al. (2006) and Wilson et al., (2002) reported that the systemic infections have considerable morbidity among those with severely paralyzed immune system.
The most common fungal infection is oropharyngeal candidiasis, which is more prevalent among cancer patients (Akapan & Morgan, 2002). Recently infections with non-Candida albicans and rare common yeast genera, such as Pichia, Rhodotorula, and Saccharomyces, have been implicated; however, C. albicans remains the most prevalent species (Han et al., 2004 and Walsh et al., 2004).
Yeast identification is of great importance for targeting proper treatments (Ramani et al., 1998), since different species yield different antifungal response (Pfaller et al., 2003). Conventional methods for yeast identification may be difficult and inconclusive (Reiss et al., 1998) especially for less common yeast. Thus more rapid and accurate molecular methods have been developed, among which ITS sequence analysis is found to be more accurate for species delineation (Chen et al., 2001 and Iwen et al., 2002).
The ITS region is located between the highly conserved genes coding for 18S and 28S rRNA. The ITS region includes two none coding regions ITS1 and ITS2, which are separated by the highly conserved 5.8SrRNA gene (White et al., 1990). The more genetic variability of ITS1 and ITS2 regions enables better identification of closely related species other than the adjacent rRNA gene (Ciardo et al., 2006).
This study aimed to detect the prevalence of oral yeast species isolated from cancer patients by oral swab, in addition to other specimens (sputum and urine), using ITS sequence analysis, since little is known about this problem in Sudan.
This was a cross-sectional, case-control study conducted in a period between April 2013 and December 2017.
The study involved 333 cancer patients referred to Isotope and Radiation Center, Khartoum, who were seeking anticancer treatment and during a routine check-up were enrolled in this study. The participants were classified into 168 patients under chemo and/or radiotherapy treatment (study group) and 165 cancer patients prior to starting anticancer treatment (control group). Study participants included 185 females and 148 males (mean age, 48 years old).
Written informed consent and structural questionnaire (see Supplementary File 1), including demographical data, site of cancer and cancer treatments, was obtained from each patient. The participants were ensured of anonymity and that only group findings will be reported.
To reduce any possible bias matching criteria was done, which included age, sex and type of cancer. Inclusion criteria: Any patient diagnosed with any type of cancer during or before starting cancer treatment, having age equal or above 18 year old, attending Isotope and Radiation Center, Khartoum was included in this study. Exclusion criteria: Patients on antifungal therapy for past two weeks were excluded from the study.
Oral swabs were collected from each patient (333 patients) to detect the prevalence of yeast infections and colonization among the treated and non-treated patients. Urine (n=9) and sputum (n=14) samples were collected from patients under treatment (study group), who exhibited the clinical features of urinary tract infection (UTI) and/or lower respiratory tract infection (LRT), respectively. These samples were taken in order to detect whether there is any dissemination from a patient’s own oral yeast due to action of cancer treatments.
All specimens (n=356) were cultured without delay in Sabouraud’s dextrose agar plates (SDA) to which chloramphenicol (0.05g/l) was added. Then the plates were incubated at 37°C for 24–48 hours. Phenotypic identification was made using Gramʹs stain and germ tube test. Purified colonies were preserved on glycerol stock solution for molecular identification (Sherman et al., 1986).
The isolated strains were subbed from the stock solution on Sabouraud’s dextrose agar medium and DNA extraction was performed from colonies that had been incubated for 48 hrs using Guanidine Chloride method as described by Gassoum et al. (2014). Three to five colonies were washed with 5 ml phosphate buffer saline (PBS) (Sigma Aldrich) for three times. 2 ml white cell lysis buffer and 20 µl of proteinase K (10 mg/ml; iNtRON Inc, Korea) were added, vortexed and incubated at 37°C for overnight. Then 1 ml from Guanidine chloride (7M; iNtRON Inc, Korea) and 350 µl of ammonium acetate (7M; Loba Chemie, India) were added. The tubes were vortexed and incubated at 65°C in an oven for 2 hours. Then the supernatant was mixed with 2ml pre chilled chloroform (sd Fine-Chem limited, India) at 6000 RPM for 20 minutes and this was transferred into a new Falcon tube and completed to 10 ml volume with pre chilled absolute ethanol (Carlo Erba, France) and incubated overnight at -20°C for completion of DNA precipitation.
After incubation the tubes were centrifuged at 6000 RPM for 20 minutes, then the ethanol was poured off and the same step was repeated with 70% ethanol. After that the tubes were left to air dry. Finally DNA was suspended in 80 µl TE buffer (iNtRON Inc, Korea) and incubated at 4 °C until used. Nanodrop ND 1000 Spectrophotometer (NanoDrop Technologies, Inc.) was used to measure quality and quantity of DNA.
The universal fungal primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) (Macrogen Inc. Korea) were used to amplify the entire ITS rDNA region (Zimbeck et al., 2010). PCR mixture contained 5 μl pre mix (iNtRON Inc., Korea), 22μl deionized sterile water, 1 μl from each forward and reverse primer, and 1 μl of genomic DNA, which served as the DNA template in a final volume of 25 μl.
PCR cycling conditions were as follows: an initial denaturation step of 5 min at 95°C followed by 35 cycles of 45 s at 94°C, 45 s at 55°C, and 45 s at 72°C, with a final extension of 5 min at 72°C. The reactions were carried out in an ESCO thermocycler (AERIS-BG096, China).
The PCR products were analyzed on 2% agarose gels (iNtRON Inc., Korea) stained with ethidium bromide (10 ng/100 ml; Fisher Scientific, USA) and visualized under a UV transilluminator apparatus (Saratoga, CA.95070, USA Gel Documentation System) and Biodoct BDA system (Biometra, Germany).
PCR products of 90 isolates from the cases and 87 isolates from the control group were purified and commercially sequenced using forward primer ITS1 and backward primer ITS4 by Macrogen Company (Seoul, Korea).
The sequences obtained in this study were identified by searching databases using BLAST sequence analysis tool (http://www.ncbi nlm.nih.gov/BLAST/). The sequences were compared using nucleotide-nucleotide BLAST (blastn) with default setting except that sequences were not filtered for low complexity. Species were identified based on the highest similarity score (100%) with reference database sequence.
Data was analyzed using SPSS 21. Frequencies and percent were obtained for frequency tables, Chi-squared test was used for goodness of fit. The relationships between variables tested were obtained using cross tables and Chi-squared (Fisher exact) test for independence. P-value ≤0.05 was considered as significant.
The collected oral swabs (333 specimens), from 168 cancer patients with chemo and/or radiotherapy treatment (study group) and from 165 cancer patients (control group) without treatment were examined by cultural technique. Oral Candida species were isolated from 69/168 (41.1%) and 74/165 (44.8%) of patients among study and control groups, respectively.
Of the 168 study group, 23 patients with clinical symptoms of UTI and/or LRT infection had urine (n=9) and sputum (n=14) samples collected for detection of Candida species. The results showed that 2 out of 9 patients and 8 out of 14 patients were culture positive for Candida spp. (Table 1).
| Specimens | |||
|---|---|---|---|
| Growth | Sputum N (%) | Urine N (%) | Total N (%) |
| Positive | 8 (57.1) | 2 (22.2) | 10 (43.5) |
| Negative | 6 (42.9) | 7 (77.8) | 13 (56.5) |
| Total | 14 (100) | 9 (100) | 23 (100) |
Among the culture positive patients (n=69 study group; n= 74 control group), the prevalence of oral candidiasis were 33.3% and 10.8% ,while the prevalence of oral colonization were 66.7% and 89.2% among the study and control patients, respectively (Table 2).
| Symptoms | ||||
|---|---|---|---|---|
| Patients | Yes (infection) N (%) | No (colonization) N (%) | Total N (%) | P- value |
| Study | 23 (33.3) | 46 (66.7) | 69 (100) | < 0.05 |
| Control | 8 (10.8) | 66 (89.2) | 74 (100) | < 0.05 |
Among the 69 positive cases in the study group, 13 patients exhibited mixed growth of oral yeast, while among the control group 25 patients exhibited mixed growth out of 74. As a result, the numbers of total isolates collected from different specimens from the study group were 92 and 99 isolates from the control group.
PCR products of ITS polymerized region revealed different band size for different species. C. albicans exhibited 500 bp (Figure 1). In contrast, non-C. albicans Candida and less common yeast represented different band sizes ranging from 400–750 bp (Figure 2).
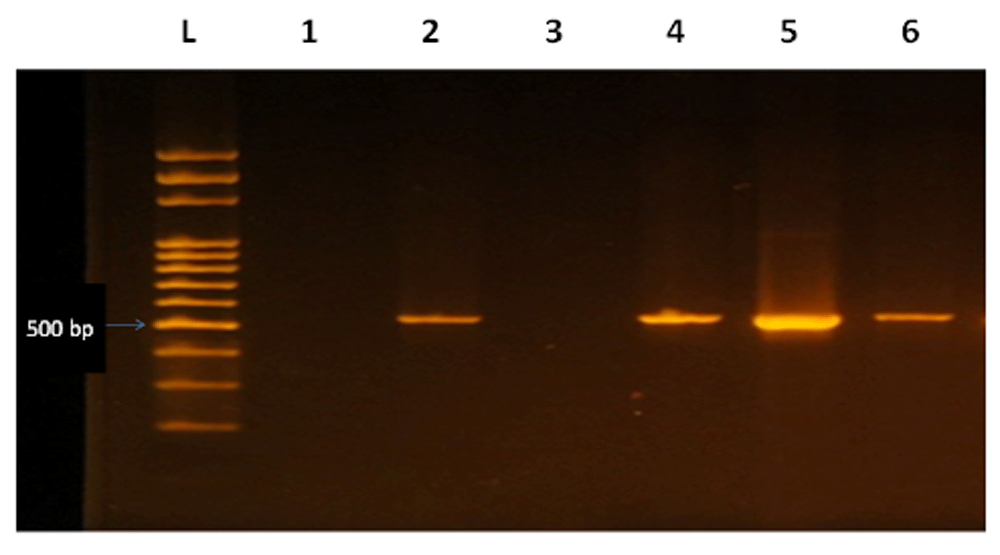
From left to right lanes: L, 100bp DNA ladder, L1 negative control, L2,4,5,6 C. albicans (500bp).
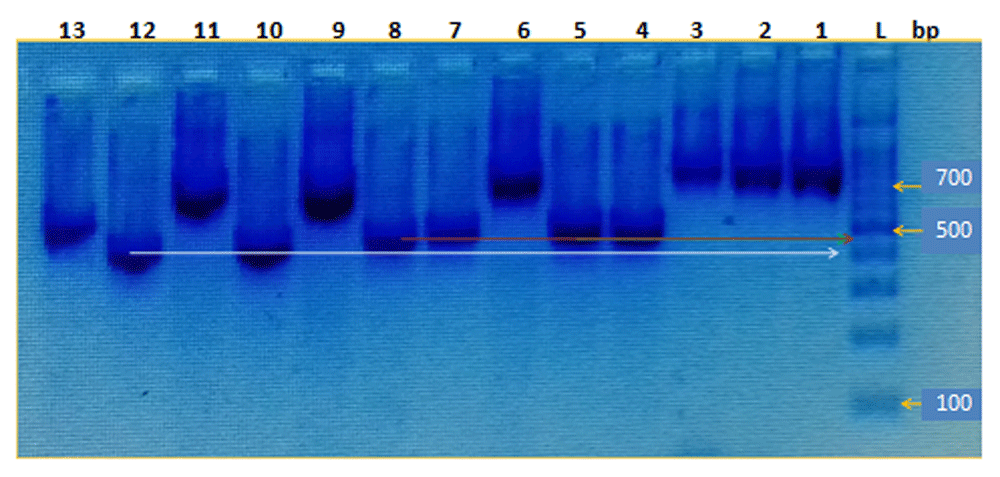
From right to left lanes: L, 100bp DNA ladder, L1,2,3,6 C. glabrata (750bp), L9,11 C. galbrata (600bp), L4,8 P. kudriavzevii (450bp), L10,12 P. kudriavzevii (400bp), L5,7 C. tropicalis (450bp), L13 L. fermentati (500bp).
DNA sequencing was performed for isolated yeast species (only 90 isolates from 67 out of 69 positive patients in study group and 87 isolates from 66 out of 74 positive patients in the control group). The result of the BLAST sequence analysis for species identification is shown in Table 3.
The distribution of oral mixed species among the 67 study and 66 control groups is shown in Table 4.
Table 4 shows the distribution of oral mixed species among the (67) study and (66) control groups. It was found that although some patients represented different colonial morphology of mixed oral isolated yeast, they exhibited the same species on sequence identification.
The identified species were classified according to presence or absence of symptoms of oral candidiasis among the study and control group to detect their association with oral infection and colonization as in Figure 3. This showed that C. albicans was the most common organism associated with oral infection (14 out of 21) and colonization (28 out of 46) among the study group. In the control group it had the same percentage (2/8; 25%) as C. tropicalis and mixed C. glabrata among oral symptomatic patients, whereas it was found to be the most common cause of colonization (21/58; 36.2%).
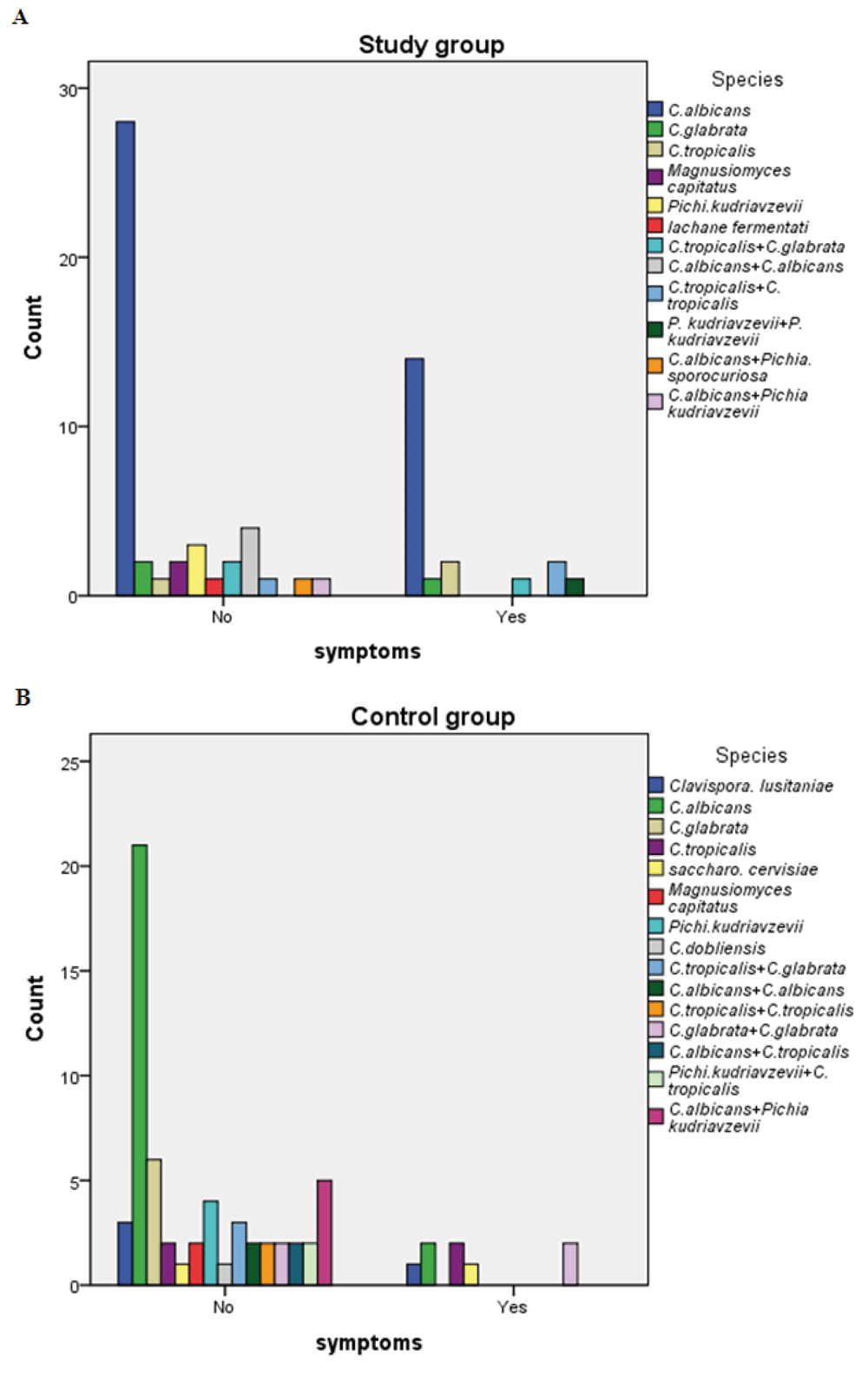
Frequency of yeast species associated with oral Candida infection (symptoms) and colonization (no symptoms) among the (A) study group and (B) control group.
Identification of sputum isolates showed that C. albicans was isolated from both oral and sputum specimens from four patients, while three patients exhibited C. albicans from sputum specimens only (oral swabs were negative), and C. tropicalis was isolated from sputum specimen only from one patient. Regarding urine specimens, C. albicans was isolated from both oral and urine samples from one patient in contrast to mixed infections (Pichia kudriavzevii and Pichia kudriavzevii) that were isolated from oral swabs. C. glabrata was also isolated from the urine of another patient.
In the present study it was found that oral Candida isolates were obtained from 69 (41.1%) patients who were under treatment with chemo and/or radiotherapy treatments and from 74 (44.8%) patients without cancer treatment. Xu et al. (2013) found that oral infection was prevalent in 46% (391/850) of all cancer patients, while another study reported the incidence of oral candidiasis ranging from 7 to 52% in cancer patients on chemotherapy and/or radiotherapy (Lone et al., 2014)
The present study revealed that 8 out of 14 patients were positive for sputum culture among the study group (Table 1). Mohammed et al. (2016) and Ungureanu et al. (2016) reported slightly lower percentages of 30.50% and 33.75%, respectively, in comparison to this study (57.15%). This may be due to differences in study population, since cytotoxic and immunosuppressive therapies promote dissemination of Candida spp. through induction of cytopenias and/or immune cell dysfunction (Blot et al., 2008; Cruciani & Serpelloni, 2008). Also this was expressed in the isolation of 22.2% of Candida species from urine culture of the treated patients among this study, which is relatively similar to Nigar et al. (2016), who found out of 64 culture positive clinical specimens Candida species were identified from 18.75% urine specimens.
Among the study and control groups, the present study found that oral colonization was significantly (p<0.05) more common than oral Candida infection (66.7% vs 33.3% and 89.2% vs 10.8%, in both groups, respectively). Similar results have been reported by Lone et al. (2014), who found the total colonization to be prevalent in 50% and oral candidiasis in 30% of all cancer patients. In contrast, oral Candida infection was more common (p<0.05) among the study group compared with the control group (33.3% vs 10.8%). A systematic review carried out by Lalla et al. (2010) reported that for all cancer treatments, the weighed prevalence of clinical oral fungal infection was found to be 7.5% pretreatment and 39.1% during treatment. This may be due to chemotherapeutic agents and therapeutic radiation that disrupts the mucosal banner of the mouth, leading to severe oral mucositis, gingivitis, and oral candidiasis. Blasting results of ITS sequence analysis in the present study revealed C. albicans was the most prevalent organism in the two groups (65.6% and 39.1%, study and control groups, respectively). These results are in agreement with previous reports (Ismet et al., 2016 and Aldossary et al., 2016) that studied the prevalence of oral Candida spp and demonstrated that C. albicans was the most prevalent organism.
The majority of oral infections are due to C. albicans but non-albicans strains, such as C. glabrata and C. tropicalis, have increasingly been implicated in causing disease (Bagg et al., 2003). Similar findings were observed in this study C. tropicalis was found to be the second most common isolate among the study group (14.4%) followed equally (7.8%) by C.glabrata and P. kudriavzevii, whereas among the control group C.glabrata was the second most common isolate (19.5%) followed by C. tropicalis (18.4%).
It was found that P. kudriavzevii (anamorph of Candida krusei) represented 12.6% among the control group, and P. sporocuriosa was isolated once from the study group only. In addition, C. lusitaniae (teleomorph of Candida lusitaniae) represented 4.6%, and C. dubliniensis represented 1.1% among the control group only (Table 3). These findings are in agreement with Pfaller et al. (2010), who performed a recent ten-year analysis of the worldwide distribution of non-albicans Candida species, and indicated that C. glabrata remains the most common non-albicans species and that C. parapsilosis, C. tropicalis, and C. krusei are also frequently isolated.
Less common yeast species were also detected in small numbers. L. fermentati appeared only once and M. capitatus twice among the study group, while among the control group the later along with S. cervisiae were detected among two patients (2.2%). These findings were in agreement with a study done by Han et al., 2004 and Walsh et al., 2004, who reported that, recently, infections caused by less common yeast species such as Pichia, Rhodotorula, Trichosporon, and Saccharomyces spp. and other rarely encountered species have been reported.
Table 4 shows the presence of identified mixed oral yeast species among the study and control groups. Similarly de Sousa et al. (2016) found the presence of more than one yeast among orogastric cancer patients.
Regarding distribution of species according to presence or absence of symptoms of oral candidiasis, it was found that C. albicans was the most common isolate associated with infection 14/21 (66.7%) and colonization 28/46 (60.9%) among the study group. While among non-albicans Candida, the prevalence of C. tropicalis was 9.5% vs. 2.2% and C. glabrata was 4.8% vs. 4.3% among the oral symptomatic and non-symptomatic study patients, respectively (Figure 3A). Similarly Lone et al., (2014) observed C. albicans to be the most common species (74.39% vs. 65.4%) causing colonization and candidiasis in cancer patients, respectively. Whereas they found C. glabrata was the second most common species followed by C. tropicalis and C. parapsilosis to cause colonization as well as candidiasis in cancer patients. This may be due to geographical region variation.
In contrast, among the control group C. albicans was detected as the same percentage (2/8; 25%) as C. tropicalis and the mixed C. glabrata among oral symptomatic patients, whereas it represents the most common cause of colonization (21/58; 36.2%) followed by C. glabrata (Figure 3B). This may be due to small number of symptomatic control patients due to the absence of any cancer treatment since it is a predisposing factor for initiation of oral infection by the colonized organisms, mainly by C. albicans, which is the most virulent organism.
Among the isolated Candida species from sputum and urine specimens, C. albicans was isolated from both oral and sputum specimens from four patients and from both oral and urine from one patient. These patients may have gained the infection or colonization in these organs from their own oral colonized Candida species. Other studies reported that colonized Candida can invade the underlying mucosa and enter the blood stream leading onto disseminated disease with considerable morbidity and mortality if not treated promptly (Lalla et al., 2010 and Shokohi et al., 2011).
The present study demonstrates that cancer patients were highly colonized with oral yeast species. C. albicans was the most common isolate associated with oral infection and colonization among the treated cancer patients. In contrast with control group it occupied a higher percent among the colonized species only. As the control group were not under cancer treatment, this lead to oral infection and disseminations to other organs. So early detection and identification of colonized yeast is of great value especially among patients undergoing cancer treatments.
Although C. albicans was the most prevalent species, other non-albicans Candida and rarely encountered yeast were also isolated. This indicates that use of proper and accurate molecular methods for yeast identification, especially for unusual yeast species, and prior antifungal treatment as required in cancer patients.
Dataset 1: Demographical data of cancer patients. Sheet 1: For case group (168 cancer patients under treatments); Sheet 2: For control group (165 cancer patients); Sheet 3: Pictures demonstrated the colonial morphology for different isolated yeast species. DOI, 10.5256/f1000research.14019.d199685 (Nagla et al., 2018).
Sequences from the patients are available on GenBank under accession numbers: MH037201 to MH037237, MH019244 to MH019255, MH016295-MH016371, MH061321-MH061334, MH084778-MH084790, MH016252 to MH016274 and MH104613.
The project was financially supported by Ministry of Higher Education and Scientific Research, Sudan.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Mycology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 11 Apr 18 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)