Keywords
contact tracing, outbreaks, R
This article is included in the RPackage gateway.
contact tracing, outbreaks, R
In order to study, prepare for, and intervene against disease outbreaks, infectious disease modellers and public health professionals need an extensive data analysis toolbox. Disease outbreak analytics involve a wide range of tasks that need to be linked together, from data collection and curation to exploratory analyses, and more advanced modelling techniques used for incidence forecasting1,2 or to predict the impact of specific interventions3,4. Recent outbreak responses suggest that for such analyses to be as informative as possible, they need to rely on a wealth of available data, including timing of symptoms, characterisation of key delay distributions (e.g. incubation period, serial interval), and data on contacts between patients5–8.
The latter type of data is particularly important for outbreak analysis, not only because contacts between patients are useful for unravelling the drivers of an epidemic9,10 , but also because identifying new cases early can reduce ongoing transmission via contact tracing, i.e. follow-up of individuals who reported contacts with known cases11,12. However, curating contact data and linking them to existing line lists of cases is often challenging, and tools for storing, handling, and visualising contact data are often missing13,14.
Here, we introduce epicontacts, an R15 package providing a suite of tools aimed at merging line lists and contact data, and providing basic functionality for handling, visualising and analysing epidemiological contact data. Maintained as part of the R Epidemics Consortium (RECON), the package is integrated into an ecosystem of tools for outbreak response using the R language.
epicontacts is released as an open-source R package. A stable release is available for Windows, Mac and Linux operating systems via the CRAN repository. The latest development version of the package is available through the RECON Github organization. At minimum users must have R installed. No other system dependencies are required.
# install from CRAN install.packages("epicontacts") # install from Github install.packages("devtools") devtools::install_github("reconhub/epicontacts")
# load and attach the package library(epicontacts)
Data handling. epicontacts includes a novel data structure to accommodate line list and contact list datasets in a single object. This object is constructed with the make_epiconctacts() function and includes attributes from the original datasets. Once combined, these are mapped internally in a graph paradigm as nodes and edges. The epicontacts data structure also includes a logical attribute for whether or not this resulting network is directed.
The package takes advantage of R’s generic functions, which call specific methods depending on the class of an object. This is implemented in several places, including the summary.epicontacts() and print.epicontacts() methods, both of which are respectively called when the summary() or print() functions are used on an epicontacts object. The package does not include built-in data, as exemplary contact and line list datasets are available in the outbreaks package16.
# install the outbreaks package for data install.packages("outbreaks")
# load the outbreaks package library(outbreaks) # construct an epicontacts object x <- make_epicontacts(linelist=mers_korea_2015[[1]], contacts = mers_korea_2015[[2]], directed=TRUE) # print the object x
##
## /// Epidemiological Contacts //
##
## // class: epicontacts
## // 162 cases in linelist; 98 contacts; directed
##
## // linelist
##
## # A tibble: 162 x 15
## id age age_class sex place_infect reporting_ctry loc_hosp
## * <chr> <int> <chr> <fct> <fct> <fct> <fct>
## 1 SK_1 68 60-69 M Middle East South Korea Pyeongtaek St˜
## 2 SK_2 63 60-69 F Outside Midd˜ South Korea Pyeongtaek St˜
## 3 SK_3 76 70-79 M Outside Midd˜ South Korea Pyeongtaek St˜
## 4 SK_4 46 40-49 F Outside Midd˜ South Korea Pyeongtaek St˜
## 5 SK_5 50 50-59 M Outside Midd˜ South Korea 365 Yeollin C˜
## 6 SK_6 71 70-79 M Outside Midd˜ South Korea Pyeongtaek St˜
## 7 SK_7 28 20-29 F Outside Midd˜ South Korea Pyeongtaek St˜
## 8 SK_8 46 40-49 F Outside Midd˜ South Korea Seoul Clinic,˜
## 9 SK_9 56 50-59 M Outside Midd˜ South Korea Pyeongtaek St˜
## 10 SK_10 44 40-49 M Outside Midd˜ China Pyeongtaek St˜
## # ... with 152 more rows, and 8 more variables: dt_onset <date>, dt_report
## # <date>, week_report <fct>, dt_start_exp <date>, dt_end_exp <date>,
## # dt_diag <date>, outcome <fct>, dt_death <date>
##
## // contacts
##
## # A tibble: 98 x 4
## from to exposure diff_dt_onset
## <chr> <chr> <fct> <int>
## 1 SK_14 SK_113 Emergency room 10
## 2 SK_14 SK_116 Emergency room 13
## 3 SK_14 SK_41 Emergency room 14
## 4 SK_14 SK_112 Emergency room 14
## 5 SK_14 SK_100 Emergency room 15
## 6 SK_14 SK_114 Emergency room 15
## 7 SK_14 SK_136 Emergency room 15
## 8 SK_14 SK_47 Emergency room 16
## 9 SK_14 SK_110 Emergency room 16
## 10 SK_14 SK_122 Emergency room 16
## # ... with 88 more rows# view a summary of the object summary(x)
##
## /// Overview //
## // number of unique IDs in linelist: 162
## // number of unique IDs in contacts: 97
## // number of unique IDs in both: 97
## // number of contacts: 98
## // contacts with both cases in linelist: 100 %
##
## /// Degrees of the network //
## // in-degree summary:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.00 1.00 1.00 1.01 1.00 3.00
##
## // out-degree summary:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.00 0.00 0.00 1.01 0.00 38.00
##
## // in and out degree summary:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.000 1.000 1.000 2.021 1.000 39.000
##
## /// Attributes //
## // attributes in linelist:
## age age_class sex place_infect reporting_ctry loc_hosp dt_onset dt_report week_report dt_start_exp dt_end_exp dt_diag outcome dt_death
##
## // attributes in contacts:
## exposure diff_dt_onsetData visualisation. epicontacts implements two interactive network visualisation packages: visNetwork and threejs17,18. These frameworks provide R interfaces to the vis.js and three.js JavaScript libraries respectively. Their functionality is incorporated in the generic plot() method (Figure 1) for an epicontacts object, which can be toggled between either with the “type” parameter. Alternatively, the visNetwork interactivity is accessible via vis_epicontacts() (Figure 2), and threejs through graph3D() (Figure 3). Each function has a series of arguments that can also be passed through plot(). Both share a color palette, and users can specify node, edge and background colors. However, vis_epicontacts() includes a specification for “node_shape” by a line list attribute as well as a customization of that shape with an icon from the Font Awesome icon library. The principal distinction between the two is that graph3D() is a three-dimensional visualisation, allowing users to rotate clusters of nodes to better inspect their relationships.
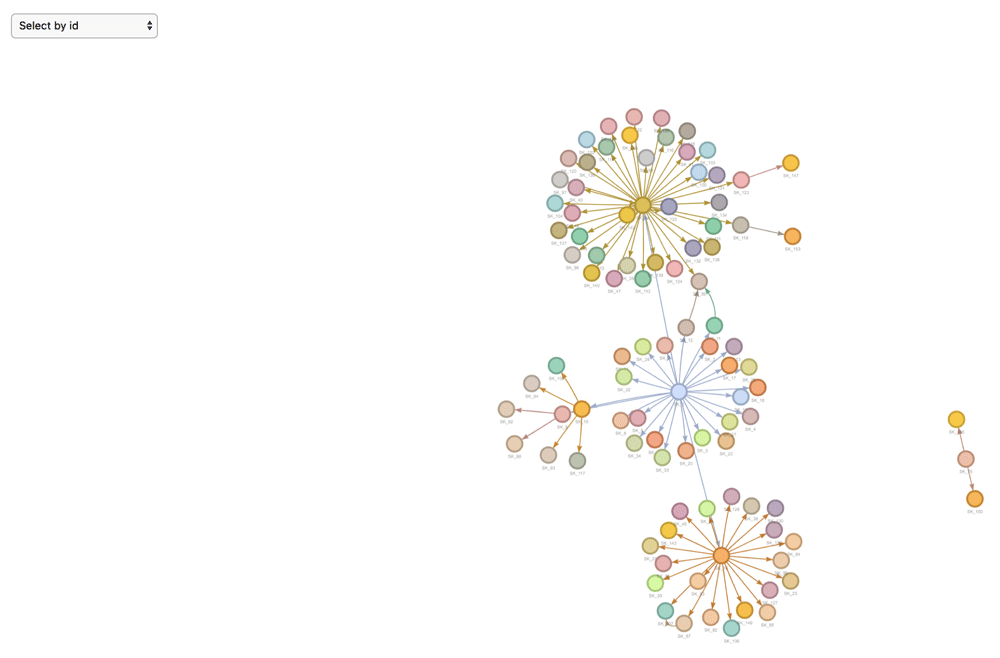

plot(x)
vis_epicontacts(x, node_shape = "sex", shapes = c(F = "female", M = "male"), edge_label = "exposure")
graph3D(x, bg_col = "black")
Data analysis. Subsetting is a typical preliminary step in data analysis. epicontacts leverages a customized subset method to filter line list or contacts based on values of particular attributes from nodes, edges or both. If users are interested in returning only contacts that appear in the line list (or vice versa), the thin() function implements such logic.
# subset for males subset(x, node_attribute = list("sex" = "M")) # subset for exposure in emergency room subset(x, edge_attribute = list("exposure" = "Emergency room")) # subset for males who survived and were exposed in emergency room subset(x, node_attribute = list("sex" = "M", "outcome" = "Alive"), edge_attribute = list("exposure" = "Emergency room")) thin(x, "contacts") thin(x, "linelist")
For analysis of pairwise contact between individuals, the get_pairwise() feature searches the line list based on the specified attribute. If the given column is a numeric or date object, the function will return a vector containing the difference of the values of the corresponding “from” and “to” contacts. This can be particularly useful, for example, if the line list includes the date of onset of each case. The subtracted value of the contacts would approximate the serial interval for the outbreak19. For factors, character vectors and other non-numeric attributes, the default behavior is to print the associated line list attribute for each pair of contacts. The function includes a further parameter to pass an arbitrary function to process the specified attributes. In the case of a character vector, this can be helpful for tabulating information about different contact pairings with table().
# find interval between date onset in cases get_pairwise(x, "dt_onset") # find pairs of age category contacts get_pairwise(x, "age_class") # tabulate the pairs of age category contacts get_pairwise(x, "age_class", f = table)
Those interested in using epicontacts should have a line list of cases as well as a record of contacts between individuals. Both datasets must be enumerated in tabular format with rows and columns. At minimum the line list requires one column with a unique identifier for every case. The contact list needs two columns for the source and destination of each pair of contacts. The datasets can include arbitrary features of case or contact beyond these columns. Once loaded into R and stored as data.frame objects, these datasets can be passed to the make_epicontacts() function (see ‘Methods’ section for more detail). For an example of data prepared in this format, users can refer to the outbreaks R package.
# load the outbreaks package library(outbreaks) # example simulated ebola data # line list str(ebola_sim$linelist) ## ‘data.frame’: 5888 obs. of 9 variables: ## $ case_id : chr "d1fafd" "53371b" "f5c3d8" "6c286a" ... ## $ generation : int 0 1 1 2 2 0 3 3 2 3 ... ## $ date_of_infection : Date, format: NA "2014-04-09" ... ## $ date_of_onset : Date, format: "2014-04-07" "2014-04-15" ... ## $ date_of_hospitalisation: Date, format: "2014-04-17" "2014-04-20" ... ## $ date_of_outcome : Date, format: "2014-04-19" NA ... ## $ outcome : Factor w/ 2 levels "Death","Recover": NA NA 2 1 2 NA 2 1 2 1 ... ## $ gender : Factor w/ 2 levels "f","m": 1 2 1 1 1 1 1 1 2 2 ... ## $ hospital : Factor w/ 11 levels "Connaught Hopital",..: 4 2 7 NA 7 NA 2 9 7 11 ...
# contact list str(ebola_sim$contacts)
## ’data.frame’: 3800 obs. of 3 variables: ## $ infector: chr "d1fafd" "cac51e" "f5c3d8" "0f58c4" ... ## $ case_id : chr "53371b" "f5c3d8" "0f58c4" "881bd4" ... ## $ source : Factor w/ 2 levels "funeral","other": 2 1 2 2 2 1 2 2 2 2 ...
# example middle east respiratory syndrome data # line list str(mers_korea_2015$linelist)
## ’data.frame’: 162 obs. of 15 variables: ## $ id : chr "SK_1" "SK_2" "SK_3" "SK_4" ... ## $ age : int 68 63 76 46 50 71 28 46 56 44 ... ## $ age_class : chr "60-69" "60-69" "70-79" "40-49" ... ## $ sex : Factor w/ 2 levels "F","M": 2 1 2 1 2 2 1 1 2 2 ... ## $ place_infect : Factor w/ 2 levels "Middle East",..: 1 2 2 2 2 2 2 2 2 2 ... ## $ reporting_ctry: Factor w/ 2 levels "China","South Korea": 2 2 2 2 2 2 2 2 2 1 ... ## $ loc_hosp : Factor w/ 13 levels "365 Yeollin Clinic, Seoul",..: 10 10 10 10 1 10 10 13 10 10 ... ## $ dt_onset : Date, format: "2015-05-11" "2015-05-18" ... ## $ dt_report : Date, format: "2015-05-19" "2015-05-20" ... ## $ week_report : Factor w/ 5 levels "2015_21","2015_22",..: 1 1 1 2 2 2 2 2 2 2 ... ## $ dt_start_exp : Date, format: "2015-04-18" "2015-05-15" ... ## $ dt_end_exp : Date, format: "2015-05-04" "2015-05-20" ... ## $ dt_diag : Date, format: "2015-05-20" "2015-05-20" ... ## $ outcome : Factor w/ 2 levels "Alive","Dead": 1 1 2 1 1 2 1 1 1 1 ... ## $ dt_death : Date, format: NA NA ...
# contact list str(mers_korea_2015$contacts)
## ’data.frame’: 98 obs. of 4 variables: ## $ from : chr "SK_14" "SK_14" "SK_14" "SK_14" ... ## $ to : chr "SK_113" "SK_116" "SK_41" "SK_112" ... ## $ exposure : Factor w/ 5 levels "Contact with HCW",..: 2 2 2 2 2 2 2 2 2 2 ... ## $ diff_dt_onset: int 10 13 14 14 15 15 15 16 16 16 ...
While there are software packages available for epidemiological contact visualisation and analysis, none aim to accommodate line list and contact data as purposively as epicontacts20–22. Furthermore, this package strives to solve a problem of plotting dense graphs by implementing interactive network visualisation tools. A static plot of a network with many nodes and edges may be difficult to interpret. However, by rotating or hovering over an epicontacts visualisation, a user may better understand the data.
The maintainers of epicontacts anticipate new features and functionality. Future development could involve performance optimization for visualising large networks, as generating these interactive plots is resource intensive. Additionally, attention may be directed towards inclusion of alternative visualisation methods.
epicontacts provides a unified interface for processing, visualising and analyzing disease outbreak data in the R language. The package and its source are freely available on CRAN and GitHub. By developing functionality with line list and contact list data in mind, the authors aim to enable more efficient epidemiological outbreak analyses.
Software available from: https://CRAN.R-project.org/package=epicontacts
Source code available from: https://github.com/reconhub/epicontacts
Archived source code as at time of publication: https://zenodo.org/record/121099323
Software license: GPL 2
The authors would like to thank all of the organizers and participants of the Hackout3 event held in Berkeley, California June 20–24, 2016. In particular, the authors acknowledge the support of the following organizations: MRC Centre for Outbreak Analysis, and Modelling at Imperial College London, the NIHR’s Modelling Methodology Health Protection Research Unit at Imperial College London, and the Berkeley Institute for Data Science.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 11 Oct 18 |
read | ||
|
Version 1 10 May 18 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)